Pituitary adenomas are uncommon, difficult to diagnose tumors whose heterogeneity and low incidence complicate large-scale studies. The Molecular Registry of Pituitary Adenomas (REMAH) was promoted by the Andalusian Society of Endocrinology and Nutrition (SAEN) in 2008 as a cooperative clinical-basic multicenter strategy aimed at improving diagnosis and treatment of pituitary adenomas by combining clinical, pathological, and molecular information. In 2010, the Spanish Society of Endocrinology and Nutrition (SEEN) extended this project to national level and established 6 nodes with common protocols and methods for sample and clinical data collection, molecular analysis, and data recording in a common registry (www.remahnacional.com). The registry combines clinical data with molecular phenotyping of the resected pituitary adenoma using quantitative real-time PCR of expression of 26 genes: pituitary hormones (GH-PRL-LH-FSH-PRL-ACTH-CGA), receptors (somatostatin, dopamine, GHRH, GnRH, CRH, arginine–vasopressin, ghrelin), other markers (Ki67, PTTG1), and control genes. Until 2015, molecular information has been collected from 704 adenomas, out of 1179 patients registered. This strategy allows for comparative and relational analysis between the molecular profile of the different types of adenoma and the clinical phenotype of patients, which may provide a better understanding of the condition and potentially help in treatment selection. The REMAH is therefore a unique multicenter, interdisciplinary network founded on a shared database that provides a far-reaching translational approach for management of pituitary adenomas, and paves the way for the conduct of combined clinical-basic innovative studies on large patient samples.
Los adenomas hipofisarios son tumores infrecuentes de diagnóstico complejo, cuya heterogeneidad y baja incidencia dificultan estudios a gran escala. El Registro Molecular de Adenomas Hipofisarios (REMAH) nació en 2008 en el seno de la Sociedad Andaluza de Endocrinología y Nutrición (SAEN), como estrategia de cooperación clínico-básica y multicéntrica, para mejorar el diagnóstico y tratamiento de tumores hipofisarios mediante la combinación de información clínica, anatomopatológica y molecular. En 2010, la Sociedad Española de Endocrinología y Nutrición (SEEN) lo extendió a nivel nacional, estableciendo 6 nodos con protocolos y métodos comunes de recogida de muestras y datos clínicos, análisis molecular y anotación en un mismo registro (www.remahnacional.com). El registro combina datos clínicos con el fenotipado molecular del adenoma intervenido, mediante PCR cuantitativa en tiempo real de la expresión de 26 genes: hormonas hipofisarias (GH-PRL-LH-FSH-PRL-ACTH-CGA), receptores (somatostatina, dopamina, GHRH, GnRH, CRH, arginina-vasopresina, ghrelina), otros marcadores (Ki67, PTTG1) y genes de control. Hasta 2015 se ha obtenido información molecular de 704 adenomas, de los 1.179 pacientes registrados. Esta estrategia permite abordar análisis comparativos y relacionales entre el perfil molecular de los distintos tipos de adenomas y el fenotipo clínico del paciente, lo que puede ofrecer un mejor conocimiento de la enfermedad y, potencialmente, ayudar en la selección del tratamiento. El REMAH constituye una red única, multicéntrica e interdisciplinar, cimentada en una base de datos compartida, que aporta un enfoque traslacional de gran proyección potencial para el manejo de los adenomas hipofisarios y abre el camino para estudios conjuntos clínico-básicos innovadores con un elevado número de pacientes.
Pituitary tumors represent a complex and significant challenge for current medicine, requiring the joint, coordinated approach of the departments of endocrinology, neurosurgery, pathology, and radiology, as well as other support disciplines. In this setting, information provided in recent decades by cellular and molecular biology on pituitary adenomas has made a substantial contribution to advances in our understanding of this group of diseases and in the design of new diagnostic and prognostic markers, as well as new therapeutic targets and strategies. There is, however, a significant gap between molecular studies of basic research and the clinical management of patients with pituitary tumors which is preventing a more rapid and effective impact of scientific findings on patient health. In order to reduce this gap, an initiative of collaboration between basic research and clinical practice for the study of pituitary tumors was launched. Its starting point was the creation of a registry of tumors that includes, in addition to the clinical data of patients, the molecular analysis of the pituitary tumor specimen taken at surgery, with both the expression levels of a set of genes selected in accordance with their potential diagnostic value and the possible clinical value of the information they provide being assessed. This article describes the general lines which made it possible to launch this initiative, called the Molecular Registry of Pituitary Adenomas (REMAH), as well as the main characteristics of molecular phenotyping.
It is now accepted that pituitary adenomas result from the clonal expansion of a specific type of adenohypophyseal cell1–3; the resulting clinical syndromes derive from the production of one or several hormones, or from local growth.4,5 Thus, the main types of pituitary adenomas include somatotropinomas producing growth hormone (GH),6 prolactinomas producing prolactin (PRL),7 corticotropinomas producing adrenocorticotropic hormone (ACTH),8 thyrotropinomas producing thyrotropin (TSH),9 gonadotropinomas producing luteinizing hormone (LH) or follicle-stimulating hormone (FSH),10 and non-functioning pituitary adenomas,11 mostly derived from the gonadotropic lineage, producing alpha subunit glycoproteins (CGA).
The classification of pituitary adenomas appears at first sight to be relatively simple and generic, which may suggest that the diagnosis of these tumors is simple, but this is not the case. In fact, pituitary cells with the mixed secretion of two or more hormones, or different cell populations, causing combined syndromes frequently appear.6,7,9–13 Silent adenomas (different from the above mentioned non-functioning tumors) with little or no hormone expression, and tumors with cyclic functionality, all of them very difficult to detect, may also occur. A detailed pathological analysis is therefore essential to better diagnose these conditions.14,15 All of this, in turn, emphasizes the need to improve the currently available tools in order to improve clinical diagnosis and the phenotyping of pituitary adenomas.
The hormone secretion of pituitary cells and many of their trophic functions (survival, proliferation, and the maintenance of specific gene expression patterns) are primarily regulated by hypophysotropic hypothalamic hormones, a set of neuroendocrine peptides having either stimulatory effects, such as GHRH, GnRH, CRH, and TRH, or inhibitory effects, such as somatostatin. There are also primary non-peptide hypothalamic regulators, such as dopamine, a potent inhibitor.4,5,16–18 All these hormones act upon their target cells through specific membrane receptors, modulating signal transduction pathways and proteins in the secretory pathway. Although the presence of these receptors is altered in many adenomas,4,5,16–21 most tumors express to a greater or lesser extent receptors of the inhibitory hormones somatostatin and dopamine, which have served as targets for drugs used in the treatment of several pituitary diseases.19,22–24
In addition to the above mentioned primary regulators, there are other factors able to regulate pituitary function, such as ghrelin, whose receptor, GHSR1a, is abundantly expressed in the pituitary gland.25–29 The role of AVP and its receptors (AVPR1a, AVPR1b, and AVPR2) at pituitary level is critical for the maintenance of basal homeostasis, and in response to stress;30 in fact, recent studies have shown the key value of AVPR1b in Cushing's disease.31,32 Securin (PTTG1) is involved in cell transformation from hyperplasia to adenoma, and its presence is related to angiogenesis;33,34 the biomarker Ki67 has been used as an immunohistochemical marker of cell proliferation, although its use and actual value remain controversial.33,34 A deeper understanding of this series of molecules and their changes in pituitary adenomas may help us to better understand the disease and to take more adequate decisions regarding its treatment and monitoring. The identification in these tumors of new regulatory molecules would also make it possible to test their effect and that of new analogs in such tumors, thus increasing the spectrum of adenomas that may be treated with drugs.
Because of the heterogeneity of pituitary tumors and the importance of knowing their molecular profile, the REMAH project was started in 2008 by the work group on neuroendocrinology of the Andalusian Society of Endocrinology and Nutrition (SAEN). It was devised and developed by basic and clinical researchers to generate information and to provide endocrinologists and their neuroendocrine teams with a helpful service for the management of patients with pituitary tumors. In 2010, this project attracted the interest of the work group on neuroendocrinology of the Spanish Society of Endocrinology and Nutrition (SEEN), which decided to endorse the project from its foundation (FSEEN), with the support of Novartis as sponsor. REMAH thus found itself going nationwide and being organized into six nodes, each coordinated by a basic researcher with molecular experience and a clinical researcher experienced in the care of patients with neurosurgical pituitary disease. Researchers in charge of patient care and monitoring and those responsible for molecular phenotyping are ascribed to each node. Node distribution is as follows: Andalusia (Cordova), the Community of Madrid (Madrid), the Valencian Community (Alicante), Galicia (Santiago de Compostela), Catalonia (Barcelona), and the Basque Country (Bilbao). The molecular study performed at each node comprises the same 26 genes, whose expression levels are tested using a systematic, standardized recording and measurement method. The measurement of gene expression provides a better understanding of the disease at molecular level and has the potential to improve treatment decisions. In addition, as a complementary objective, the REMAH project aims to collect the molecular results of each pituitary tumor, together with clinical patient data, in a common registry that will provide all participating researchers with a research platform containing a significant number of cases of a disease with a low prevalence, so helping Spanish neuroendocrinology to improve its levels of international scientific competitiveness.
From the scientific viewpoint, the general objective of REMAH is to determine the expression in pituitary tumors of different genes corresponding to receptors, hormones, and other regulatory proteins in order to improve diagnosis. This allows for the additional objective of assessing the relationship of those parameters to the clinical and pathological characteristics of the patients, and thus of analyzing their potential role in the development and pathological response of pituitary adenomas. Specifically, the testing of proteins related to the control of hormone secretion, survival, or cell apoptosis that may affect these conditions, makes it conceivable that the potential value and relevance of molecules and signals tested as treatment targets may be established, and that early diagnostic markers of these diseases can be obtained. These data are thus expected to be of value for both the clinical and basic researchers involved in the study.
To achieve this general objective, the following specific objectives were established: 1) To assess the expression of pituitary hormones and the main hypothalamic hormone receptors, with priority being given to those related to the disease evaluated: somatostatin (sst1, sst2, sst3, and sst5), dopamine (DRD1, DRD2T, DRD2L, DRD4, and DRD5), GHRH (GHRH-R), CRH (CRH-R1), AVP (AVPR1b), and ghrelin (GHSR1a). 2) To study the expression of other proteins that regulate secretion or cell proliferation and death, which may be used as markers or new treatment targets, PTTG1 and Ki67. 3) To determine the potential relationship of the expression levels of these receptors and molecules of interest with patient response to specific drug treatments for the different types of pituitary tumors, and with the clinical characteristics and course of those patients. More specifically, the purpose of this publication is to detail how REMAH works, its methodology, and the tumor samples collected to date.
Patients and methodsRecruitment of patients. Informed consentThe REMAH study has been conducted in accordance with the principles of the Declaration of Helsinki and approved by the ethics committees of participating hospitals. Informed consent was obtained from each patient before study entry.
Methodology and design of the REMAH study, consisting of four phases:
Case preparationDiagnosisThe following factors were considered necessary:
- •
Functional identification of the tumor and its specific clinical characteristics.
- •
Evaluation of the type of response to stimuli, enhancers, or suppressors of hormone secretion in functioning tumors.
- •
Identification of tumor sequelae affecting the rest of the hypothalamic-pituitary structure.
- •
Assessment of preoperative drug response (somatostatin analogs, adrenal steroidogenesis inhibitors, etc.), the type of drug, duration and its effect on hormone secretion (if applicable).
- •
Replacement therapy for associated deficiencies before surgery.
- •
Assessment of the need for the use of steroid therapy during surgery.
- •
Blood sample collection before drug treatment or surgery.
- •
Decision and programming of surgical procedures.
- •
Hospital admission.
- •
The collection of clinical history data from the case report form attached to the REMAH protocol, included in the corresponding database. Specifically, fasting levels of the following substances are measured in circulating blood: glucose (mg/dL), HbA1c (%), cholesterol (mg/dL), LDL (mg/dL), HDL (mg/dL), basal GH (ng/mL), PRL (mU/mL), ACTH (pg/mL), LH (mU/mL), FSH (mU/mL), TSH (μU/mL), CGA (mU/mL), IGF-I (ng/mL), free T4 (ng/L), and cortisol (μg/dL). Report after surgery: cure after the procedure and pharmacological control in the event of no cure. Imaging data: adenoma size (<1cm, microadenoma or >1cm, macroadenoma), extrasellar, suprasellar, and infrasellar extension, characteristics in T1 and T2 MRI sequences.
- •
It should be noted that the registry allows for (and has among its future objectives) detailed patient monitoring after surgery or under medical treatment, including blood sample collection at 6 and 12 months.
Sampling is performed using a standardized procedure. Each participating center applies its own coordination system to ensure the collection of adequate samples from all tumors operated on at the hospital, according to the requirements of the agreed procedure. Procedure coordination and supervision depends on the researchers in charge of both sample collection and the corresponding clinical data.
- 1.
The surgical specimen is processed as an “intraoperative” sample for pathology, with measures being taken to prevent any delay in collection. In neurosurgery, the adenoma specimen is immediately placed in cold physiological saline, never in formalin, and no more than 30min should elapse from removal to processing.
- 2.
The specimen is assessed by the pathology department, which provides, after taking the portion required for diagnosis, a representative fragment that is transferred to a cryotube with RNAlater stabilizing solution (Life Technologies, Carlsbad, CA, USA); if the fragment is greater than 0.5cm in size, it is divided to allow for the penetration of the stabilizing substance. The cryotube is identified with a REMAH code, generated in the computer application/registry, which is linked to the corresponding code of the patient's clinical history and to the report of the endocrinology department. The cryotube with the sample is stored at 4°C until transferred to the reference node selected.
- 3.
The adenoma sample is shipped to the reference node. The tumor sample is shipped in the cryotube itself, either under refrigerated conditions or at room temperature.
Parameters tested during the course of the overall project are detailed below:
- •
Demographic data: sex and age.
- •
Molecular expression levels, measured by real time quantitative PCR (qPCR): GH, PRL, POMC, β subunits of luteinizing, follicle-stimulating, and thyroid-stimulating hormones (LHB, FSHB, and TSHB respectively), α subunit of glycoproteins (CGA), sst1, sst2, sst3, sst5, DRD1, DRD2T, DRD2L, DRD4, DRD5, GnRH-R, GHRH-R, CRH-R1, GHSR1a, AVPR1b, Ki67, PTTG1, and β-actin control genes (ACTB), glyceraldehide-3-phosphate dehydrogenase (GAPDH), and hypoxanthine–guanine phosphoribosyltransferase (HPRT).
- •
Biochemical levels in blood, report after surgery, and indicated imaging data.
Tumor tissue fragments undergo standardized processing procedures for gene expression assessment. Depending on the type of tumor, the characteristics of the disease under study, and the size of the available sample, priorities are established for the genes to be tested based on the needs for clinical information concerning the disease. RNA extraction from the sample is performed as previously described,29 using AllPrep RNA/DNA/Prot supplemented with RNase-Free DNase Set (#80004 and #79254 respectively, Qiagen, Limburg, the Netherlands) following the manufacturer's instructions. The tumor specimen is homogenized in cold using RLT buffer supplemented with beta-mercapto-ethanol, using a pellet pestle tissue grinder with a cordless drive unit (#749515-0000 and #749540-0000 respectively, Kontes, Sigma–Aldrich, Madrid, Spain), after which RNA isolation is performed using the column system included in the kit. RNA is then eluted in 30–50μL of DEPC-treated water, depending on the sample size. Reverse transcription of RNA is performed using a RevertAid FirstStrand cDNA synthesis kit (#K1622, Fermentas, Hanover, MD, USA) according to the manufacturer's instructions, using random hexamers. Specifically, the use of 0.5μg of RNA by reaction, with a double reaction (1μg of RNA) to obtain a final volume of 40μL of copy DNA, which is stored at −20°C until measurement by real time quantitative PCR (qPCR) is considered optimum.
Primer selectionAll pairs of primers have been designed using the genomic sequences of GenBank (National Center for Biotechnology Information) and Primer3 software,35,36 as previously reported,17 using the following criteria: a) the difference in binding temperature between both primers does not exceed 0.2°C, b) primers that generate primer dimers have been excluded, and 3) they amplify a product by between 100 and 200 base pairs. Primer sequences were verified using BLAST (National Center for Biotechnology Information) to avoid potential homology to other sequences. Primer sequence, expected sizes, and GenBank numbers are shown in Table 1.
Primer sequence, expected sizes, and GenBank numbers for the genes studied.
| Gene | GenBank no. | Sequence | Size | |||
|---|---|---|---|---|---|---|
| Sense (bp) | Antisense (bp) | |||||
| ACTB | NM_001101 | ACTCTTCCAGCCTTCCTTCCT | 21 | CAGTGATCTCCTTCTGCATCCT | 22 | 176 |
| GAPDH | NM_002046 | AATCCCATCACCATCTTCCA | 20 | AAATGAGCCCCAGCCTTC | 18 | 122 |
| HPRT | BT019350 | CTGAGGATTTGGAAAGGGTGT | 21 | TAATCCAGCAGGTCAGCAAAG | 21 | 157 |
| GH | NM_000515 | GACCTAGAGGAAGGCATCCAAA | 22 | AGCAGCCCGTAGTTCTTGAGTAG | 23 | 143 |
| PRL | BC015850 | CCTTCGAGACCTGTTTGACC | 20 | ATCTGTTGGGCTTGCTCCTT | 20 | 183 |
| POMC | BC065832 | CCCTACAGGATGGAGCACTT | 20 | CGTTCTTGATGATGGCGTTT | 20 | 127 |
| LHB | NM_000894 | GCCTCCTCTTCCTCTAAAGACC | 22 | GCGGATTGAGAAGCCTTTATT | 21 | 104 |
| FSHB | NM_000510 | TTGGTGTGCTGGCTACTGCT | 20 | GGGCACTCTCACTGTTTCGT | 20 | 115 |
| TSHB | NM_000549.3 | ATTGCCTAACCATCAACACCAC | 22 | AAACATCCTGGGACAGAGCATA | 22 | 102 |
| CGA | NM_000735.2 | GCAAAAAGCCCAGAGAAAGG | 20 | ATCAGGAGCGGAATGGAGAA | 20 | 107 |
| sst1 | NM_001049 | CACATTTCTCATGGGCTTCCT | 21 | ACAAACACCATCACCACCATC | 21 | 165 |
| sst2 | NM_001050 | GGCATGTTTGACTTTGTGGTG | 22 | GTCTCATTCAGCCGGGATTT | 20 | 185 |
| sst3 | NM_001051 | TGCCTTCTTTGGGCTCTACTT | 22 | ATCCTCCTCCTCAGTCTTCTCC | 22 | 190 |
| sst5 | NM_001053 | CTGGTGTTTGCGGGATGTT | 19 | GAAGCTCTGGCGGAAGTTGT | 20 | 183 |
| DRD1 | AF498961 | GACCACCACAGGTAATGGAAAG | 22 | AAGAAAGGTAGCCAACAGCACA | 22 | 141 |
| DRD2T | NM_016574 | CGAGCATCCTGAACTTGTGTG | 21 | GCGTTATTGAGTCCGAAGAGG | 21 | 172 |
| DRD2L | NM_000795 | CTCCTCCATCGTCTCCTTCT | 20 | CGGTGCAGAGTTTCATGTCC | 20 | 188 |
| DRD4 | L12398 | GACGCCCTTCTTCGTGGT | 18 | GACAGTGTAGATGACGGGGTTG | 22 | 130 |
| DRD5 | AY136750 | CTGGGCTAACTCCTCACTCAAC | 22 | ATTGCTGATGTTCACCGTCTC | 21 | 130 |
| GnRH-R | AY392011.1 | TGCCTCTTCATCATCCCTCTT | 21 | AGTCTTCAGCCGTGCTCTTG | 20 | 144 |
| GHRH-R | NM_000823 | TCACCATCCTGGTTGCTCTC | 20 | GCAGCATCCTTCAGGAACAC | 20 | 112 |
| CRH-R1 | NM_004382 | TTTTCAACATCGTCCGCATC | 20 | GGGATTGACGAAGAACAGCA | 20 | 143 |
| GHSR1a | NM_198407.2 | TGAAAATGCTGGCTGTAGTGG | 21 | AGGACAAAGGACACGAGGTTG | 21 | 148 |
| AVPR1b | NM_000707.3 | ACAAGAATGCCCCTGATGAA | 20 | GGCTGTTGAAGCCCATGTAG | 20 | 111 |
| KI67 | NM_002417 | GACATCCGTATCCAGCTTCCT | 21 | GCCGTACAGGCTCATCAATAAC | 22 | 139 |
| PTTG1 | NM_004219.2 | GGCTGTTAAGACCTGCAATAATC | 23 | TTCAGCCCATCCTTAGCAAC | 20 | 101 |
To verify primer specificity, as previously described,17 each pair of primers was used in conventional PCR (DreamTaq DNA Polymerase, Thermo Scientific, Wilmington, NC, USA) to amplify copy DNA generated by reverse transcription from human pituitary tissue. The thermal profile consists of a 10/min step at 95°C followed by 35 cycles of one minute at 95°C, one minute at 60–64°C and one minute at 72°C. Products were run on agarose gel stained with ethidium bromide to confirm the presence of a single band of the expected size and the absence of primer dimers. An aliquot of the PCR product was purified using the AccuPrep Gel Purification Kit (K3035, Bioneer, Alameda, CA, USA), and sequencing was performed to confirm primer specificity. The conventional PCR product was used to construct standard curves for quantitative PCR. To confirm primer efficiency and construct standard curves, the first assay with qPCR was performed with a 1:2 dilution of reverse transcription being made, in which optimum efficiency is shown by a difference of one Ct between both dilutions. Details of qPCR reagents are given in the next section. The concentration of the purified product was measured using the PicoGreen DNA Quantification kit (Molecular Probes, Eugene, OR, USA), and PCR products were sequentially diluted, to obtain standards of 101, 102, 103, 104, 105, and 106 copies of transcript by microliter. One microliter of each point in the curve is amplified by qPCR, and a standard curve relating Ct and the number of copies is thus generated. The R2 values generated from the straight line ranged from 0.997 to 1.003; primer pairs with efficiency ranging from 90% to 110% were accepted; 100% efficiency indicates that all transcripts are amplified in each cycle. If all validation parameters were adequate, the pair of primers was selected, and their reaction conditions were used to amplify the copy DNA of tumor samples.
Real time quantitative PCR (qPCR)Gene expression is measured using real time quantitative PCR. MasterMix Brilliant III ultrafast SYBRGreen QPCR (#600882, Agilent, La Jolla, CA, USA), 96-well plates and lids (#B70501 and #B79791B respectively, Bioplastic, Landgraaf, the Netherlands) were used. The reaction is as follows: 10μL of MasterMix, 1μL of copy DNA, 150nM of each primer, and up to 20μL DEPC-treated water. The thermal profile consists of a 3-min step at 95°C, 40 cycles of 20s at 95°C followed by 20s at 60°C, and a final dissociation step, one minute at 95°C, 30s at 55°C, and 30s at 95°C. The result of qPCR is a Ct value which, when entered into the standard curve equation, generates an expression value in the number of copies. Extrapolation is performed using a scatter plot with dilution exponents 1, 2, 3, 4, 5, and 6 and the corresponding Ct values obtained in qPCR, thus generating a straight line and its subsequent equation of the straight line: y=mx+q, where m is the slope of the straight line, which should approximate to 3.3, the difference in Ct between the different dilutions, and q is the intersection point with axis y, and the Ct value of the sample is the value of x; thus, the replacement yields its value in the number of copies.
Correlation of expression with clinical and pathological characteristics, behavior, and tumor progression.
Results achieved in the above sections are grouped by type of tumor and treatments and tests conducted and, based on this, the medical records of patients are reviewed to assess their main characteristics.
Statistical analysisThe different nature of the study variables requires detailed data analysis. Komogorov–Smirnov tests were performed to determine the similarity of the different sets with a normal distribution. A Student's t test was used to compare variables with parametric distributions, while a Mann–Whitney test was conducted for non-parametric variables. For more than two groups, Kruskal–Wallis tests were performed. Correlation analysis was performed using Spearman correlation coefficient.
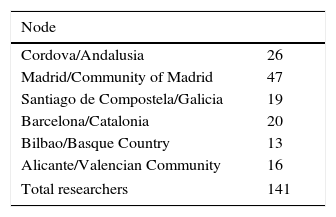
ResultsCases registeredFrom its inception the REMAH project has recruited a growing number of researchers with authorized access to the database. At the end of the first phase of the project, there were 141 researchers, whose geographical distribution is detailed in Table 2.
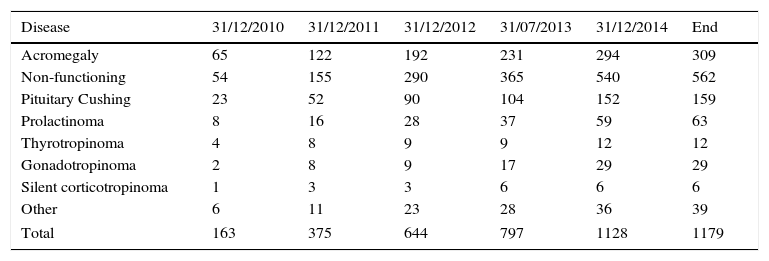
The cumulative registry in the REMAH database in this first phase consisted of 1179 entries corresponding to pituitary disease recorded with REMAH code (Table 3). Non-functioning pituitary adenoma was especially common (562), followed by acromegaly (309), and Cushing's disease (159). Overall, the distribution of prevalence of the different diseases was similar in the different nodes.
Cumulative registry of samples and prevalence of the corresponding diseases according to their clinical phenotyping. By year, from 2010 to 2015.
| Disease | 31/12/2010 | 31/12/2011 | 31/12/2012 | 31/07/2013 | 31/12/2014 | End |
|---|---|---|---|---|---|---|
| Acromegaly | 65 | 122 | 192 | 231 | 294 | 309 |
| Non-functioning | 54 | 155 | 290 | 365 | 540 | 562 |
| Pituitary Cushing | 23 | 52 | 90 | 104 | 152 | 159 |
| Prolactinoma | 8 | 16 | 28 | 37 | 59 | 63 |
| Thyrotropinoma | 4 | 8 | 9 | 9 | 12 | 12 |
| Gonadotropinoma | 2 | 8 | 9 | 17 | 29 | 29 |
| Silent corticotropinoma | 1 | 3 | 3 | 6 | 6 | 6 |
| Other | 6 | 11 | 23 | 28 | 36 | 39 |
| Total | 163 | 375 | 644 | 797 | 1128 | 1179 |
Other: refers to other pituitary tumors and lesions that cannot be ascribed to the other categories or which have not been categorized separately because of their low incidence; these include pituitary apoplexy, silent gonadotropinoma, chordoma, oncociytoma, and plurihormonal adenoma. Craniopharyngioma and ectopic Cushing's disease have also been included in this section because although they are not pituitary adenomas, they are closely related to them.
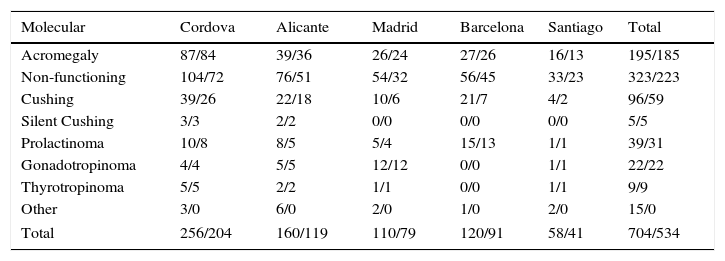
Among the total sample of 1179 cases registered, molecular data were recorded for 704 cases, corresponding to the Cordova, Alicante, Barcelona, Madrid, and Santiago de Compostela nodes. Table 4 shows the distribution of tumors registered with molecular information by type and node where they are registered, and the molecular data which were considered initially valid for statistical analysis, corresponding to 534 cases (total/validated). Cases of craniopharyngioma and ectopic ACTH-secreting tumors have also been included in this registry because of their association with pituitary disease.
Distribution of samples registered with available/validated molecular phenotype by type of tumor and registry node.
| Molecular | Cordova | Alicante | Madrid | Barcelona | Santiago | Total |
|---|---|---|---|---|---|---|
| Acromegaly | 87/84 | 39/36 | 26/24 | 27/26 | 16/13 | 195/185 |
| Non-functioning | 104/72 | 76/51 | 54/32 | 56/45 | 33/23 | 323/223 |
| Cushing | 39/26 | 22/18 | 10/6 | 21/7 | 4/2 | 96/59 |
| Silent Cushing | 3/3 | 2/2 | 0/0 | 0/0 | 0/0 | 5/5 |
| Prolactinoma | 10/8 | 8/5 | 5/4 | 15/13 | 1/1 | 39/31 |
| Gonadotropinoma | 4/4 | 5/5 | 12/12 | 0/0 | 1/1 | 22/22 |
| Thyrotropinoma | 5/5 | 2/2 | 1/1 | 0/0 | 1/1 | 9/9 |
| Other | 3/0 | 6/0 | 2/0 | 1/0 | 2/0 | 15/0 |
| Total | 256/204 | 160/119 | 110/79 | 120/91 | 58/41 | 704/534 |
Other: see Table 3.
Assessment of the stability of the three control genes initially measured in the study, ACTB, GAPDH and HPRT, showed that HPRT expression is markedly more stable than the other two. For this reason, it has been decided to use data adjusted for HPRT to compare expression levels and correlations with biochemical levels.
Somatotropinoma causing acromegalyOf the 195 somatotropinomas registered, the molecular profiles of 185 tumors were validated. Basic demographic data (Table 5) show that among the patients in this study, acromegaly is more common in females. Age at surgery is significantly younger in males (p=0.005). In both sexes, a vast majority of tumors (more than 80%) are macroadenomas.
Non-functioning pituitary adenomasThe most common tumors in the study were non-functioning pituitary adenomas. This was to be expected given their greater incidence and prevalence in the general population as compared to all other adenomas. In this first phase of REMAH, molecular analysis of 223 of the 326 non-functioning adenomas registered was started (Table 6). Adenomas with a profile more consistent with that of non-functioning adenoma were selected, and those which despite being clinically diagnosed as non-functioning showed a less evident molecular profile that could correspond to silent tumors or asymptomatic stages of a disease of another nature were proposed for more detailed subsequent study. The results found in the subgroup selected suggested a higher incidence of non-functioning adenomas in males, in whom age at surgery also tended to be older than in females (p=0.052). As expected, almost all tumors were macroadenomas in both sexes, as surgery is not indicated for non-functioning microadenomas.
Basic demographic data of non-functioning adenomas registered.
| Non-functioning | No. (%) | Age [median (min–max)] | Macroadenoma (%) |
|---|---|---|---|
| Male | 187 (57) | 61 (16–81) | 126/126 (100) |
| Female | 139 (43) | 55 (19–87) | 99/102 (97) |
| Total | 323a | 58 (16–87) | 225/228 (99) |
Samples recorded in REMAH have made it possible to start the molecular analysis of 59 of the 96 corticotropinomas causing Cushing's disease. Only 61% of the adenomas recorded were considered valid, because in many cases the molecular analysis revealed that the sample tested was not a corticotroph adenoma, but a fragment of pituitary gland with no tumor, with marked expression of GH and PRL, followed by POMC and all other adenohypophyseal hormones: this is the hormone expression profile typical of the normal pituitary gland.
In the sample analyzed (Table 7), almost two thirds of which consisted of microadenomas, the incidence was much higher in females than in males, while no significant age differences were found between the sexes.
Lactrotroph adenoma causing hyperprolactinemiaProlactinoma, causing hyperprolactinemia, is the fourth leading tumor in the series registered. Molecular analysis was performed in 31 of the 39 prolactinomas registered (Table 8). As this type of tumor is initially treated with dopamine agonists, which often make it possible to control and even resolve the disease, prolactinomas requiring surgery are most often macroadenomas, which in this series were somewhat more common in males.
DiscussionThe national REMAH project has successfully completed its first phase with the implementation of a standardized working protocol, sample collection, patient registry, and a first demographic analysis of the population registered.
From the practical viewpoint, the results collected suggest:
- 1.
The project has the capacity to recruit a substantial number (141) of clinical and basic researchers for patient collection and the testing of samples, as well as for their joint registry and study.
- 2.
It can develop a combined clinical–molecular database accessed by Internet to serve as the basis for current and future development of the project. It can already be used to support diagnosis, and further improvement will allow for its faster and more dynamic use for this purpose as well as for the future development of many different projects.
- 3.
It may be stated with confidence that the high number of patients and samples registered, their variety and types, together with the molecular characterization of tissue and detailed clinical study of cases, represent an unprecedented achievement in the research and study of pituitary tumor disease at both national and international level.
- 4.
The simple descriptive analysis of the demographic data of the cases analyzed and of the molecular profiles evaluated for the main types of pituitary adenomas has demonstrated the substantial value of the information collected both overall and, what will most likely be more important, in each individual patient.
- 5.
Future work to make the most of the data recorded and the measurements taken, and to maximize the value of the REMAH, is complex and necessarily cumbersome, but also of a quite extraordinary interest.
The results obtained to date, together with the achievement of the main milestones and the objectives initially established, allow us to state that the launching and implementation of this project have reached an adequate level of development and completion. Therefore, what has been achieved so far justifies our proposing the continuity of the designed strategy in a second phase that includes any improvements deemed appropriate in terms of the review and re-structuring of the selected genes to be measured and their clinical parameters, and also the design and implementation of new actions derived from the findings already made and, particularly, from the results obtained, which represent a product of substantial value due to the information contained in the registries made and their potential as a basis for devising new studies and initiatives.
Conflicts of interestMontserrat Gilabert is an employee of Novartis Farmacéutica. The other authors state that they have no conflicts of interest.
The REMAH project has been possible thanks to the sponsorship of Novartis, Fundación FSEEN, and SAEN. The research projects of the authors themselves have also contributed. We thank all researchers, technicians, and healthcare staff who have contributed to the project, and particularly the patients and their families, for their contribution.
Nodo Madrid:
Magdalena Adrados, Hospital Universitario de La Princesa Pedro Martínez Flores, Hospital Universitario de La Princesa
Ana María Ramos Leví, Hospital Universitario de La Princesa
Miguel Sampedro-Núñez, Hospital Universitario de La Princesa
Ana Serrano-Somavilla, Hospital Universitario de La Princesa
María Paz de Miguel Novoa, Hospital Clínico San Carlos
Juan José Díez, Hospital Ramón y Cajal
Mercedes García Villanueva, Hospital Ramón y Cajal
Pedro Iglesias, Hospital Ramón y Cajal
Víctor Rodríguez Berrocal, Hospital Ramón y Cajal
Concepción Blanco Carrerra, Hospital Príncipe de Asturias
Esperanza Aguillo Gutiérrez, Hospital Clínico Zaragoza
Luciano Bances, Hospital Clínico Zaragoza
Fernando L. Calvo Gracia, Hospital Clínico Zaragoza
Fernando Comuñas, Hospital Clínico Zaragoza
Iván Quiroga López, Hospital de Talavera
Carmen Alameda Hernando, Hospital Infanta Sofía
Jesús Miguel Pérez Luis, Hospital Universitario de Tenerife
Rogelio García Centeno, Hospital Gregorio Maranón
Begoña Iza, Hospital Gregorio Marañón
Álvaro Pérez Zamarrón, Hospital La Paz
José F. Alén, Hospital 12 de Octubre
María Calatayud Gutiérrez, Hospital 12 de Octubre
Igor Paredes Sansinenea, Hospital 12 de Octubre
Álvaro Otero, Hospital Clínico de Salamanca
José María Recio Córdova, Hospital Clínico de Salamanca
Pablo Sousa, Hospital Clínico de Salamanca
José Belinchón, Hospital Virgen de la Salud
María José Herguido, Hospital Virgen de la Salud
Ángel Rodríguez de Lope, Hospital Virgen de la Salud
Almudena Vicente Delgado, Hospital Virgen de la Salud
Nodo Barcelona:
Iris Crespo, Hospital de Sant Pau
Fernando Muñoz, Hospital de Sant Pau
Eugenia Resmini, Hospital de Sant Pau
Olga Roig, Hospital de Sant Pau
Alicia Santos, Hospital de Sant Pau
Pere Tresserras, Hospital de Sant Pau
Carlos del Pozo Pico, Mutua de Terrassa
Alberto Torres, Hospital de Bellvitge
Noemí Vidal, Hospital de Bellvitge
Carles Villabona, Hospital de Bellvitge
Gemma Sesmilo, Centro Médico Teknon
Guillermo Cuatrecasas Cambra, Centro Médico Teknon
Joaquim Enseñat, Hospital Clinic Barcelona
Irene Halperin, Hospital Clinic Barcelona
Gabriel Obiols, Hospital Vall d’Hebron
Cristina Carrato, Hospital Germans Trias
Isabel Salinas, Hospital Germans Trias
Roxana Zabala, Hospital Germans Trias
Laura Cecenarro, Hospital Germans Trias
Raquel Buj, Hospital Germans Trias
Mireia Jordá, Hospital Germans Trias
Cristina Hostalot, Hospital Germans Trias
Javier Ibáñez Domínguez, Hospital Universitario Son Espases
Honorato García Fernández, Hospital Universitario Son Espases
Guillermo Serra, Hospital Universitario Son Espases
Nodo Alicante:
Pedro Riesgo, Hospital de La Ribera
Rosa Cámara, Hospital La Fe
Juan Antonio Simal-Julian, Hospital La Fe
Cristina Lamas, Hospital General de Albacete
Hernán Sandoval, Hospital General de Albacete
Javier Abarca, Hospital General de Alicante
Nieves Arias Mendoza, Hospital General de Alicante
Ruth Sánchez Ortiga, Hospital General de Alicante
Irene Monjas, Hospital General de Alicante
Teresa Pedro Font, Hospital de Denia
Nodo Santiago:
Isabel Alonso Troncoso, Complejo Hospitalario Pontevedra
Pablo Fernández Catalina, Complejo Hospitalario Pontevedra
Rosa María Álvarez San Martín, Complejo Asistencial de León
María D. Ballesteros Pomar, Complejo Asistencial de León
Rocío Villar Taibo, Complejo Asistencial de León
Sihara Pérez Romero, Universidad Santiago Compostela
Eva Fernández Rodríguez, Hospital Clínico Universitario de Santiago
Alfredo García-Allut, Hospital Clínico Universitario de Santiago
Ramón Serramito, Hospital Clínico Universitario de Santiago
Alma Prieto, Hospital el Bierzo (León)
Laura Cotovad Bellas, Complejo Hospitalario Arquitecto Marcide
Jose Ignacio Vidal Pardo, Complejo Hospitalario XeralCalde
Nodo Córdoba:
Juan Antonio García Arnés, Hospital Universitario Carlos Haya
Inmaculada González-Molero, Hospital Universitario Carlos Haya
Silvia María Maraver Selfa, Hospital Virgen de la Victoria
Francisco J. Tinahones Madueno, Hospital Virgen de la Victoria
María Rosa Alhambra Expósito, Hospital Universitario Reina Sofía
Paloma Moreno Moreno, Hospital Universitario Reina Sofía
Andrés de la Riva, Hospital Universitario Reina Sofía
Elena Torres Vela, Hospital Universitario San Cecilio
Miguel Ángel Japón, Hospital Virgen del Rocío
María Elena Dios Fuentes, Hospital Virgen del Rocío
Natividad González Rivera, Hospital de la Macarena
Tomas Martín Hernández, Hospital de la Macarena
Inmaculada Gavilán Villarejo, Hospital Universitario Puerta del Mar Cádiz
José Gregorio Oliva García, Hospital Universitario Nuestra Señora de la Candelaria
Judith López Fernández, Hospital Universitario de Canarias
Alberto Moreno Carazo, Complejo Hospitalario de Jaén
These two authors contributed equally to this study and are considered as coprime authors.
Please cite this article as: Luque RM, Ibáñez-Costa A, Sánchez-Tejada L, Rivero-Cortés E, Robledo M, Madrazo-Atutxa A, et al. El Registro Molecular de Adenomas Hipofisarios (REMAH): una apuesta de futuro de la Endocrinología española por la medicina individualizada y la investigación traslacional. Endocrinol Nutr. 2016;63:274–284.