To assess a new immunochromatography (ICT) test that detects glutamate dehydrogenase (GDH) antigen and Clostridium difficile toxin A/B simultaneously, and to propose an algorithm for the diagnosis of C. difficile infection (CDI) based on this test.
MethodsWe analysed 970 stool samples. Discrepant results between GDH and toxin A/B were resolved using toxigenic culture as the reference.
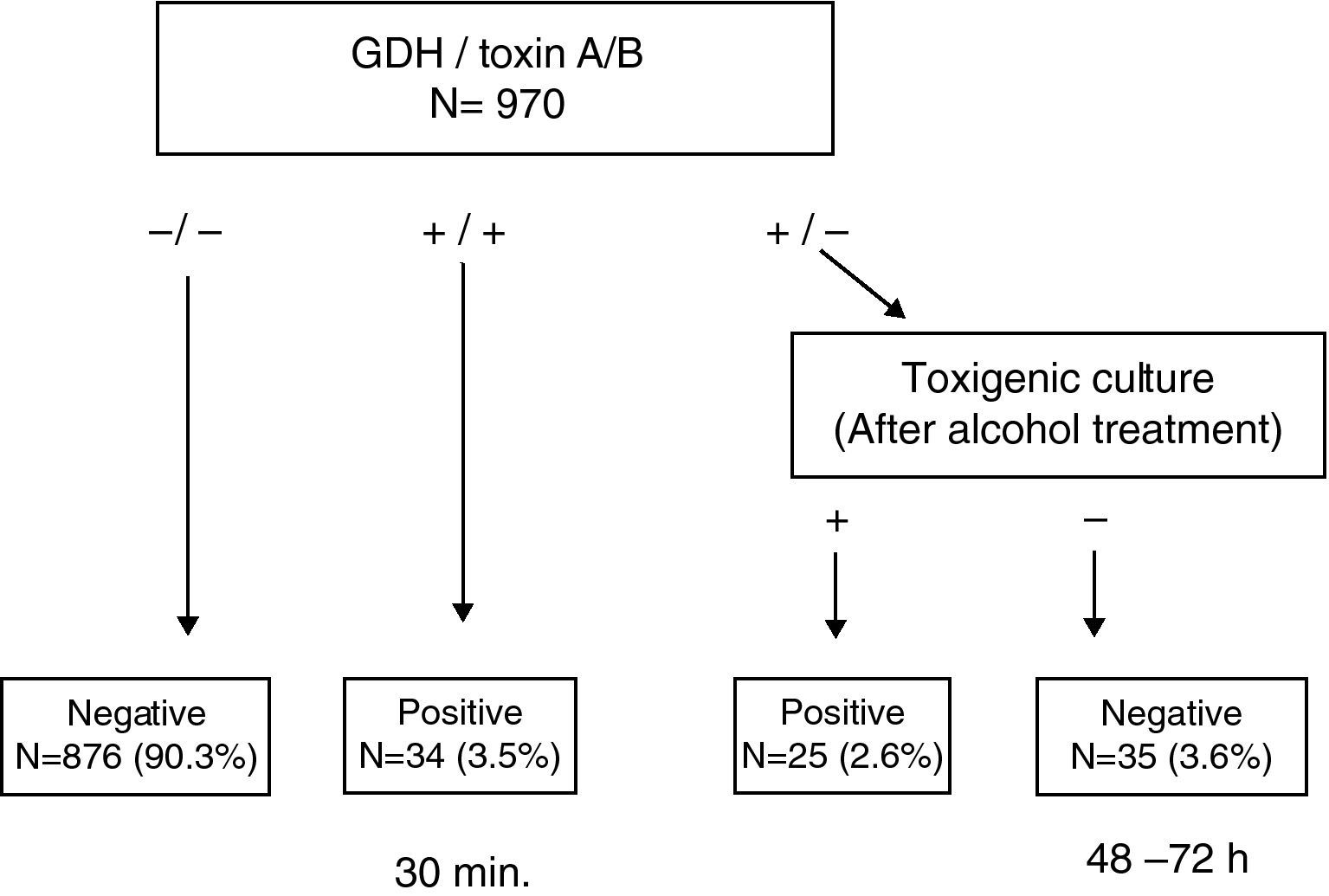
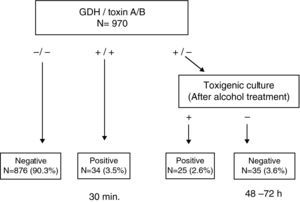
ResultsThis test enabled us to obtain a conclusive result in <30min in 93.8% of the samples. Among the discrepant results (GDH (+)/Toxin A/B (−)), 41.7% (25/60) were found to be toxigenic C. difficile by toxigenic culture.
ConclusionThis test has a high sensitivity and specificity for the diagnosis of CDI.
Evaluar una nueva prueba inmunocromatográfica que detecta el antígeno glutamato deshidrogenasa (GDH) y la toxina A/B de Clostridium difficile simultáneamente y proponer un algoritmo para el diagnóstico de infección por C. difficile (ICD) basado en esta prueba.
MétodosSe analizaron 970 muestras. Las discrepancias entre GDH y toxina A/B se resolvieron utilizando el cultivo toxigénico como método de referencia.
ResultadosEsta prueba permitió obtener el resultado del 93,8% de las muestras en <30 minutos. El 41,7% (25/60) de las muestras discrepantes (GDH (+)/Toxina A/B (−)) fueron C. difficile toxigénicos, mediante cultivo toxigénico.
ConclusiónEsta prueba es sensible y específica para el diagnóstico de ICD.
Clostridium difficile is the most common cause of nosocomial diarrhoea.1 In particular, this microorganism causes illness in individuals infected with toxin-producing strains.2–4 Typically, the diagnosis of C. difficile infection (CDI) is based on clinical history and diarrhoea in combination with in vitro laboratory tests.5 Rapid and accurate diagnosis of CDI is essential for the management of patients and institutions.1
Currently, there are several methods available for diagnosing CDI including the rapid detection of toxin A/B or glutamate dehydrogenase (GDH) antigen by enzyme immunoassay (EIA) or immunochromatographic test (ICT) devices, tissue culture neutralisation (CTN), toxigenic culture and polymerase chain reaction (PCR) for toxin gene detection.6 GDH is a constitutive enzyme that is produced in both toxigenic and non-toxigenic C. difficile strains and other members of the Clostridium genus and related genera similar to C. difficile.7 This test has high sensitivity and a negative predictive value, but low specificity,8 while the detection of toxin A/B in stools, although highly specific, has poor sensitivity, resulting in a considerable proportion of false negatives.9 CTN and toxigenic culture are considered the gold standard for C. difficile diagnosis, but they are tedious and time-consuming, requiring specific laboratory facilities and technical expertise.5 On the other hand, PCR for the toxin B gene (tcdB) directly from stools has high sensitivity and specificity compared to CTN and toxigenic culture. Further, depending on the PCR method, this approach is usually relatively quick, but it is expensive.9 Recently, a new test that detects GDH and toxin A/B in a single ICT device has been approved by the FDA. The aim of this study was to assess this new test using toxigenic culture as a reference method and to propose an algorithm for the routine diagnosis of CDI based on the use of this test.
MethodsThis study was conducted in 12 de Octubre University Hospital, a 1300-bed tertiary care facility that has a catchment population of approximately 550,000 residents in southern Madrid, Spain. Liquid or semisolid stool samples obtained from patients more than one year old and suspected of CDI were processed immediately or when this was not possible, were kept at 4°C or frozen at −70°C until processing. The samples were studied for detection of GDH and toxin A/B with a single ICT the Techlab® C. diff Quik Chek Complete device (Inverness Medical Innovations, Inc., Princeton, NJ, USA) according to the manufacturer's instructions and also, cultured on selective cycloserine-cefoxitin-fructose agar plates (CLO agar, bioMerieux, Marcy l‘Etoile, France) and incubated in an anaerobic chamber for 48–72h at 37°C according to standard laboratory methods. C. difficile was identified by its typical morphology (large, yellow colonies), characteristic “horse barn” odour and Gram stain. Presumptive colonies were subcultured on anaerobic agar plates and incubated for 48–72h and then direct toxin detection from culture was performed again, using the C. diff Quik Chek Complete device, with a suspension of 3–5 culture colonies in 750μl of dilution buffer. When the results were GDH positive, toxin A/B positive or negative, and culture negative, stool samples were treated with ethanol to kill non-spore flora (equal volumes of ethanol and stools mixed for 1h before inoculation) and cultured on CLO plates. A sample was considered a true negative when GDH, toxin A/B and culture were negative and a true GDH positive when a positive C. difficile culture was obtained.
ResultsA total of 970 stool samples were tested between February and October 2009. The rate of positive C. difficile culture was 9.0% (87/970), of which 67.8% (59/87) were toxigenic. Results from GDH, toxin A/B and toxigenic culture were concordant in 93.6% of samples (875/970 negatives and 33/970 positives); in the remaining 62 samples the results were discordant. Specifically, in 47 cases, the results of GDH and culture were positive, but toxin A/B negative. Direct toxin testing on the culture plates yielded a positive result in 23 (48.9%) of these cases. On the other hand, in 13 cases the discordance consisted of GDH positive, with toxin A/B and culture both negative. Ethanol treatment of these stool samples enabled the recovery of 6 (46.1%) C. difficile isolates, two of which were toxin A/B positive. In one sample, the GDH and toxin A/B results were positive, while the culture was negative and the culture after ethanol exposure also proved negative. In the last of these 62 samples, the discordance consisted of GDH and toxin A/B results being negative but the culture positive, the detection of toxin A/B directly from the culture being negative. There were no GDH negative and toxin A/B positive samples. Taking into account only the results of toxin A/B obtained using the ICT device, the rate of toxigenic C. difficile was 3.4%, detecting only 57.6% of the strains. This rate increased to 5.8% when toxin detection was performed directly from culture and up to 6.1% when GDH positive and culture negative stool samples were treated with ethanol and retested.
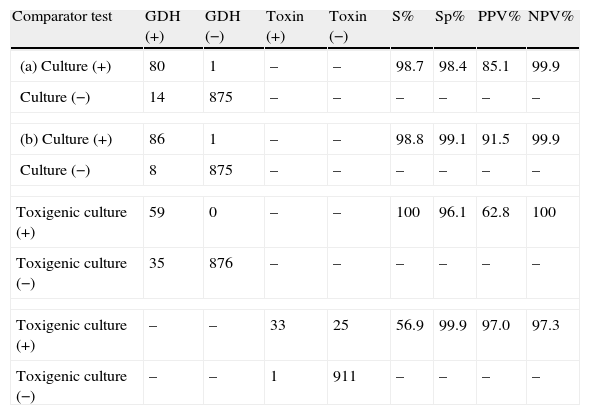
Sensitivity, specificity, positive (PPV) and negative predictive (NPV) values for GDH and toxin A/B results are given in Table 1. The sensitivity and NPV of GDH compared to toxigenic culture reached 100%, while the specificity and PPV of toxin A/B compared to toxigenic culture were 99.9% and 97.0%, respectively.
Performance of GDH and toxin A/B compared to culture and toxigenic culture.
| Comparator test | GDH (+) | GDH (−) | Toxin (+) | Toxin (−) | S% | Sp% | PPV% | NPV% |
| (a) Culture (+) | 80 | 1 | – | – | 98.7 | 98.4 | 85.1 | 99.9 |
| Culture (−) | 14 | 875 | – | – | – | – | – | – |
| (b) Culture (+) | 86 | 1 | – | – | 98.8 | 99.1 | 91.5 | 99.9 |
| Culture (−) | 8 | 875 | – | – | – | – | – | – |
| Toxigenic culture (+) | 59 | 0 | – | – | 100 | 96.1 | 62.8 | 100 |
| Toxigenic culture (−) | 35 | 876 | – | – | – | – | – | – |
| Toxigenic culture (+) | – | – | 33 | 25 | 56.9 | 99.9 | 97.0 | 97.3 |
| Toxigenic culture (−) | – | – | 1 | 911 | – | – | – | – |
(a) Before alcohol treatment, (b) after alcohol treatment, S: sensitivity, Sp: specificity, PPV: positive predictive value, NPV: negative predictive value.
Some algorithms for detecting CDI propose testing for GDH as a screening method, because of its high sensitivity (step one); and when the result is positive, testing for toxin A/B (step two). A third step is performed to resolve discrepancies, using toxigenic culture or CTN.3,6,10 The new ICT Techlab C. diff Quik Chek Complete device detects both GDH and toxin A/B and allows us to reduce such algorithms to two steps. In our study, with a large number of samples tested, in the first step, the detection of GDH had NPVs of 99.9% and 100% for all C. difficile strains and toxigenic strains respectively, and a PPV of 91.5% for all strains but of only 62.8% for toxigenic strains. The toxin A/B ICT showed very high specificity (99.9%) with only one false positive result. However, the sensitivity was poor compared to toxigenic culture (56.9%). Some other authors have reported a similarly low sensitivity,11,12 although in one study it reached 78.3%.13 It is likely that the level of expression of toxin A/B in different ribotypes of C. difficile isolates could justify these apparently discordant results.11
This simple and rapid test device allows us: (a) to rule out C. difficile without additional tests when GDH is negative (90.3%) and (b) to confirm CDI when both GDH and toxin A/B results are positive (3.5%). Notably, this step takes less than 30min. In a second step, when discrepancies appear between GDH and toxin A/B (GDH (+)/Toxin A/B (−)), the ethanol treatment of stool samples and direct toxin detection from culture enabled us to recover isolates from 41.7% (25/60) of samples that were toxin A/B negative, increasing the rate of confirmed CDI to 6.1% (Fig. 1). Although some clinical laboratories would not be able to perform this test immediately after receiving the sample and they have to delay the assay, we believe that this test device is useful, because we can obtain more than 90% of results more rapidly than by toxigenic culture. On the other hand, although the confirmation of a toxigenic effect could be obtained by tissue culture neutralisation, this is tedious, time-consuming and requires specific laboratory facilities and technical expertise.
In accordance with these results, we propose an algorithm based on this simple and rapid test device, although the performance of this may vary depending on the prevalence of CDI. To use this device no special equipment is required and it can be easily implemented in clinical laboratories. The drawback of this algorithm is that the second step involves toxigenic culture which is time-consuming. An alternative, when results between GDH and toxin A/B are discordant would be direct PCR detection from stool samples as, nowadays, there are RT-PCR systems that detect toxin B in less than 1h, this option is then relatively quick and easy to perform but is more expensive.8,11–13
Conflict of interestThe authors declare no conflict of interest.
This work was partially supported by Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008/0011).