Diabetes mellitus is a highly prevalent chronic progressive disease with complications that include diabetic-foot ulcers.
MethodsEnterococci isolated from diabetic-foot infections were identified, evaluated by macro-restriction analysis, and screened for virulence traits and antimicrobial resistance.
ResultsAll isolates were considered multidrug-resistant, cytolysin and gelatinase producers, and the majority also demonstrated the ability to produce biofilms.
ConclusionsThese results indicate the importance of enterococci in diabetic-foot infection development and persistence, especially regarding their biofilm-forming ability and resistance to clinically relevant antibiotics.
La diabetes mellitus es una enfermedad crónica progresiva de alta prevalencia cuyas complicaciones incluyen úlceras del pie.
ProcedimientoSe han identificado enterococos aislados de infecciones del pie diabético, evaluados mediante análisis de macrorrestricción y búsqueda de marcadores de virulencia y de resistencia antimicrobiana.
ResultadosTodos los aislados analizados fueron considerados multirresistentes, productores de citolisina y gelatinasa, y la mayoría fueron capaces de formar biofilms.
ConclusionesEstos resultados permiten conjeturar sobre la importancia de los enterococos en el desarrollo y la persistencia de la infección del pie diabético, fundamentalmente debido a la capacidad de formación de biofilm y de resistencia a antibióticos de relevancia clínica.
Diabetes mellitus is a serious health problem in rapid expansion worldwide.1 One the most frequent diabetes complications being the development of diabetic foot infections – DFIs, which represent a major cause of morbidity and mortality among patients. Antibiotherapy continues to be the most important approach to solve or control such infections, however, increasing bacterial resistance to a growing number of antimicrobial agents, frequently results in treatment failure. Previous reports2 point towards gram-positive cocci as the most common pathogens in DFI samples, contributing to the persistence/severity of the disease and leading to higher morbidity and mortality rates. Members of the Enterococcus genus are known to be among such bacteria. In this context, the present study aimed to evaluate the diversity, antimicrobial drug resistance, biofilm forming ability and virulence patterns of enterococci isolated from diabetic foot infections.
MethodsBacterial isolatesThe study was conducted in 4 clinical centers (2 outpatient clinics, 1 general surgery ward and 1 vascular surgery ward) in Lisbon from January 2010 to June 2010. Specimens (aspirates, biopsies and swabs) were obtained from patients with Diabetes mellitus and clinically infected foot ulcers, as advised by current clinical guidelines.3 After collection, forty-nine clinical samples were screened for the presence of Enterococcus spp. using conventional microbiological procedures.2
Molecular characterizationFollowing DNA isolation by boiling lysis, genus and species allocation were performed according to methodologies described elsewhere.4 Macrorestriction analysis by Pulsed-Field Gel Electrophoresis – PFGE – was applied as previously reported5 and data generated were analyzed using the BioNumerics 6.6 software (Applied Maths, Kortrijk, Belgium).
Virulence factorsApplication of previously described protocols6 included screening for genes coding for aggregation substance – agg, the E. faecalis antigen A – efaAfs, the enterococcal surface protein – esp, gelatinase – gelE, the cytolysin activator – cylA and plate assays for the evaluation of hemolytic and gelatinolytic phenotypes.
Evaluation of biofilm forming ability was by Fluorescent in Situ Hybridization – FISHBiofilm production was evaluated in vitro as described elsewhere.7
Antibiotic susceptibility testsSusceptibility to sixteen antimicrobial agents, representing distinct classes, was evaluated by the disk diffusion method, using previously established breakpoints of resistance.8 MICs for vancomycin were further determined using E-test.
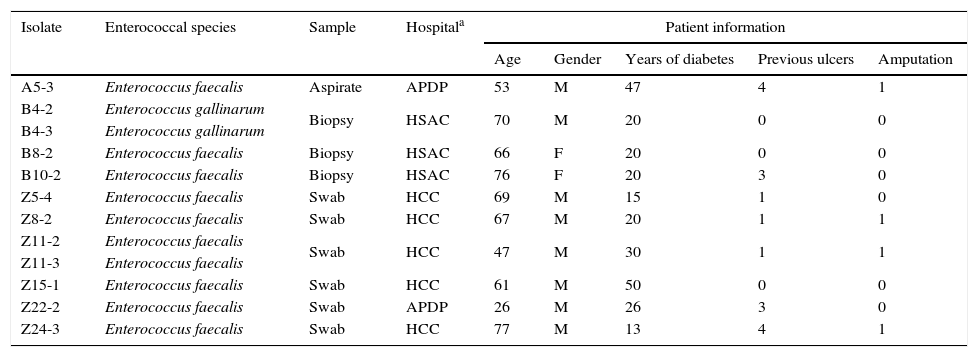
Results and discussionThe pathogenesis of foot ulceration is complex, the mortality is high and healed ulcers often recur, resulting in severe chronic foot infections. Additionally, the indiscriminate misuse and abuse of antibiotics for DFI treatment has triggered an increase in the development of multidrug-resistances, leading to serious public health issues due to treatment failure. Most DFIs have a polymicrobial etiology, enterococcal strains being part of the complex diabetic foot microbiota.1,2 The present study screened forty-nine samples from DFIs for the presence of Enterococcus spp. Twelve enterococci were recovered (see Table 1 for further details) and identified as E. faecalis (10) and E. gallinarum (2). The higher prevalence of E. faecalis among the diabetic foot ulcer enterococci corresponds to the expected, as this species is considered the most pathogenic of this genus, being commonly associated with clinical samples.9
Data regarding enterococcal isolates and DFI patients.
| Isolate | Enterococcal species | Sample | Hospitala | Patient information | ||||
|---|---|---|---|---|---|---|---|---|
| Age | Gender | Years of diabetes | Previous ulcers | Amputation | ||||
| A5-3 | Enterococcus faecalis | Aspirate | APDP | 53 | M | 47 | 4 | 1 |
| B4-2 | Enterococcus gallinarum | Biopsy | HSAC | 70 | M | 20 | 0 | 0 |
| B4-3 | Enterococcus gallinarum | |||||||
| B8-2 | Enterococcus faecalis | Biopsy | HSAC | 66 | F | 20 | 0 | 0 |
| B10-2 | Enterococcus faecalis | Biopsy | HSAC | 76 | F | 20 | 3 | 0 |
| Z5-4 | Enterococcus faecalis | Swab | HCC | 69 | M | 15 | 1 | 0 |
| Z8-2 | Enterococcus faecalis | Swab | HCC | 67 | M | 20 | 1 | 1 |
| Z11-2 | Enterococcus faecalis | Swab | HCC | 47 | M | 30 | 1 | 1 |
| Z11-3 | Enterococcus faecalis | |||||||
| Z15-1 | Enterococcus faecalis | Swab | HCC | 61 | M | 50 | 0 | 0 |
| Z22-2 | Enterococcus faecalis | Swab | APDP | 26 | M | 26 | 3 | 0 |
| Z24-3 | Enterococcus faecalis | Swab | HCC | 77 | M | 13 | 4 | 1 |
Note: The present study was approved by the Faculty of Medicine of the University of Lisbon Research Ethics Committee and the Portuguese Data Protection Authority, and written informed consent was obtained for every patient.
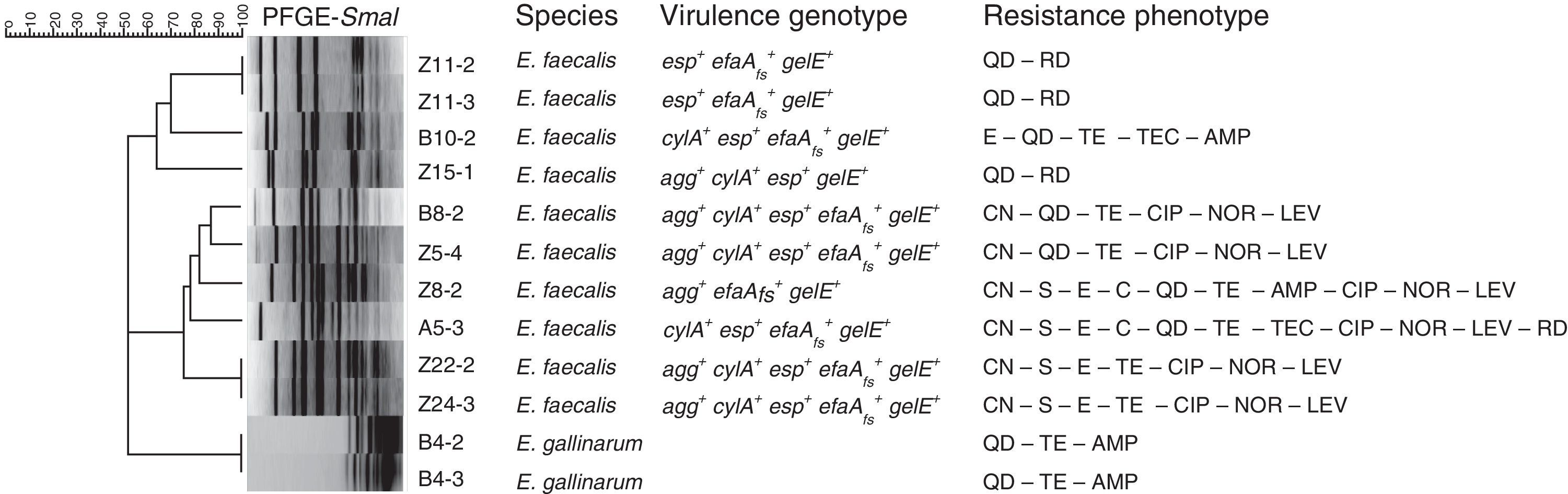
Macrorestriction analysis was the methodology chosen to assess the genomic diversity of the enterococci under study. The dendrogram built based on macrorestriction patterns (pulsotypes) allocated the enterococcal isolates into nine genomic groups (Fig. 1). Results demonstrated that no single enterococci was present in all the samples under analysis, although high similarity levels could be observed between distinct isolates, revealing their clonal relationships. Briefly, the two E. gallinarum share the same pulsotype; since they were isolated from the same patient it can be established that they are identical, or highly related. Similar conclusions can be drawn for E. faecalis Z11-2 and Z11-3 (swab samples from the same patient), as well as for Z22-2 and Z243, obtained from DFIs of patients attending distinct healthcare units, which apparently harbor the same enterococcal isolate. However, to further prove the persistence of specific enterococci in DFIs, additional sample collection should be performed over the years in the same patients.
Dendrogram based on SmaI-PFGE patterns. The BioNumerics 6.6 software (Applied Maths, Kortrijk, Belgium) was used to register macrorestriction patterns and clustering analysis was performed using Dice similarity coefficient and the unweighted-pair group method with arithmetic mean (UPGMA). Samples: A – aspirate, B – biopsy, Z – swab. Virulence determinants: agg – aggregation substance, cylA – cytolysin activator, efaAfs – cell wall adhesin, esp – cell wall-associated protein, gelE – gelatinase. Antibiotics: AMP – ampicillin, C – chloramphenicol, CIP – ciprofloxacin, QD – quinupristin-dalfopristin, E – erythromycin, CN – gentamicin-120, LEV – levofloxacin, LZD – linezolid, F – nitrofurantoin, NOR – norfloxacin, RD – rifampicin, S – streptomycin-300, TEC – teicoplanin, TE – tetracycline, VA – vancomycin.
Concerning the screening for virulence features (see Fig. 1), all DFI enterococci present hemolytic and gelatinolytic abilities and the E. faecalis DFI isolates harbor distinct virulence determinants. Since the screened virulence traits are considered among the most relevant for enterococcal pathogenicity mechanisms, often detected in clinical isolates and correlated with the persistence and severity of infection,9 these results constitute important indicators for the putative pathogenicity of the DFI enterococci under study.
Furthermore, analysis of phenotypic biofilm expression revealed 83% (10/12) of biofilm producers at 48h, with negative results being associated only with the non-E. faecalis DFI-enterococci. Due to the known importance of biofilms in the persistence of human infections, such as DFI, the biofilm forming ability demonstrated by the enterococci further demonstrates their putative contribution for the chronicity of infection.
Regarding antibiotic resistance, all isolates were simultaneously resistant to several antibiotics, representing distinct drug classes and directed towards various bacterial targets (Fig. 1). Considering as multidrug-resistant -MDR- the enterococci non-susceptible to more than 3 antibiotics representing distinct classes and bacterial targets,10 the majority of the isolates under analysis fall into the MDR category. Although vancomycin MIC determination showed that none of the isolates are resistant to this drug (MIC≤4μg/ml), the MDR status attributed to the majority of the enterococci continues to be highly relevant, especially in chronic severe infections such as DFIs, since antimicrobial resistance often results in treatment failure.
Overall, the present study demonstrated that DFI enterococci harbor virulence determinants, which are associated with biofilm-forming ability and resistance to medically important antibiotics, suggesting their contribution to the persistence and severity of diabetic foot infections. The presence of multidrug-resistant diabetic foot ulcer enterococci is of major importance also due to the possibility of transmitting those multi-drug resistances to other microorganisms sharing the same ecological niche, highly impairing the implementation of successful antibiotic treatment. Since DFI are one of the most frequent diabetes complications, which represent a major cause of morbidity and mortality among patients, further studies directed towards the evaluation of the role of enterococci, during the establishment and persistence of infection, are fundamental.
Conflict of interestThe authors declare no conflict of interest.
This study was performed on “Centro de Investigação Interdisciplinar em Sanidade Animal” (CIISA/FMV) from Faculdade de Medicina Veterinária da Universidade de Lisboa (Faculty of Veterinary Medicine, University of Lisbon), through Project PTDC/SAU-MIC/122816/2010 from “Fundação para a Ciência e Tecnologia-FCT”. Teresa Semedo-Lemsaddek is financed by Programa Ciência and Carla Mottola holds a PhD fellowship (SFRH/BD/72872/2010), both from FCT, Portugal.