To characterize a carbapenem-resistant Enterobacter cloacae complex isolate recovered from a patient from Ukraine.
MethodsThe isolate was sent to a regional reference laboratory for molecular characterization by whole genome sequencing. Susceptibility assays, carbapenemase identification, imipenem hydrolysis and clonality were performed.
ResultsThe isolate showed resistance or reduced susceptibility to all β-lactam agents tested. Genome analysis led to the identification of an NDM-1-producing E. cloacae complex strain that was assigned to a new multilocus sequence type, ST932. The blaNDM-1 enzyme was located in a conjugative IncX3 plasmid of ca. 50kb. In addition, blaCMH-3, a recently described AmpC β-lactamase sequence, which has not previously been reported in Europe, was also detected and its genetic environment was studied.
ConclusionTo our knowledge, this is the first reported case in Europe of an E. cloacae complex strain that produces both blaNDM-1 and blaCMH-3.
Caracterizar una cepa de Enterobacter cloacae complex resistente a los carbapenémicos detectado en un paciente procedente de Ucrania.
MétodosSe envió la cepa a un laboratorio regional de referencia para proceder a su caracterización molecular mediante secuenciación del genoma completo. Se realizaron estudios de susceptibilidad, identificación de carbapenemasas, hidrólisis del imipenem y de clonalidad.
ResultadosEl aislado mostró resistencia o sensibilidad reducida a todos los betalactámicos estudiados. El análisis molecular permitió la identificación de una cepa de E. cloacae complex productor de NDM-1 que se asignó a un nuevo secuenciotipo, ST932. Se localizó la enzima blaNDM-1 en plásmido conjugativo IncX3 de 50kb. Además, se detectó la enzima blaCMH-3, una nueva betalactamasa de tipo AmpC descrita recientemente pero no detectada con anterioridad en Europa, llevándose a cabo el estudio de su entorno genético.
ConclusiónSegún nuestros conocimientos, esta es la primera descripción una cepa de E. cloacae complex productora de blaNDM-1 junto con blaCMH-3 en Europa.
In recent years, Enterobacter cloacae complex has emerged as a common nosocomial pathogen with several outbreaks being reported.1E. cloacae complex naturally exhibits resistance (first and second generation cephalosporins, aminopenicillins) mediated by a chromosomal AmpC. In addition, plasmid-mediated AmpC genes have also been reported in Enterobacter spp.1 The overproduction of AmpC confers resistance to most β-lactam antibiotics, except carbapenems and fourth-generation cephalosporins. Recently, novel plasmid-mediated AmpC-type-β-lactamases (CMH) have been described in Taiwan and India.2,3
Carbapenem resistance in Enterobacter spp. has been frequently associated to the production of AmpC enzymes and/or extended-spectrum betalactamases coupled with efflux pump and porin loss.1 However, worldwide emergence and spread of carbapenemases, such as NDM, in mobile genetic elements are becoming a cause of concern.4 In Spain, NDM-1-producing Enterobacter spp. isolates have been detected but this is not the predominant carbapenemase in this genus.5–8
Thus, the aim of this study was the molecular characterization of an E. cloacae complex clinical isolate carrying blaNDM-1 and blaCMH-3.
Material and methodsIn 2016, two carbapenem-resistant E. cloacae complex strains were recovered from a patient who had a traffic accident in Ukraine and was transferred, two months later, to the Hospital Virgen de la Victoria (Málaga, Spain). The isolates were recovered from a blood culture (Ec2016198) and a rectal swab (Ec2016212) and were identified as E. cloacae complex by MALDI-TOF (Bruker Daltonics).
Antimicrobial susceptibility testing was performed by microdilution (Microscan, Beckman Coulter) and interpreted according to EUCAST (EUCAST, 2018).9 βCarba test (BioRad) was used to assess imipenem hydrolysis. The study of inhibitors (phenyl-boronic, dipicolinic acid, cloxacillin) was performed using disc diffusion (Rosco Diagnostica) according to EUCAST recommendations.10
The presence of carbapenemases (NDM/VIM/KPC/IMP/OXA-48 groups); extended-spectrum beta-lactamases (CTX-M/SHV/TEM); and plasmid-mediated AmpCs (ACC/CMY/FOX/DHA) were evaluated by PCR and DNA sequencing.11–13
Both E. cloacae complex isolates were compared by PFGE-XbaI.
Ec2016212 was selected for WGS (Illumina MiSeq Inc) and de novo assembly was performed with CLC Genomics Workbench (Qiagen). Antimicrobial resistance and plasmid characterization were determined using ResFinder and PlasmidFinder databases.
For bacterial conjugation E. coli J53 was used as recipient. For plasmid electroporation, the plasmid DNA of Ec2016212 was used to electroporate into E. coli DH10B. Several colonies of transconjugants and electroporants were tested by microdilution and plasmid transfer by PCR.
The genetic context of the blaNDM-1 gene was analyzed by SI-PFGE and Southern Blot using probes specific to blaNDM-1 and IncX3. The Whole Genome project has been deposited at GenBank under the following BioProject No. PRJNA529367 (E. cloacae isolate Ec2016212).
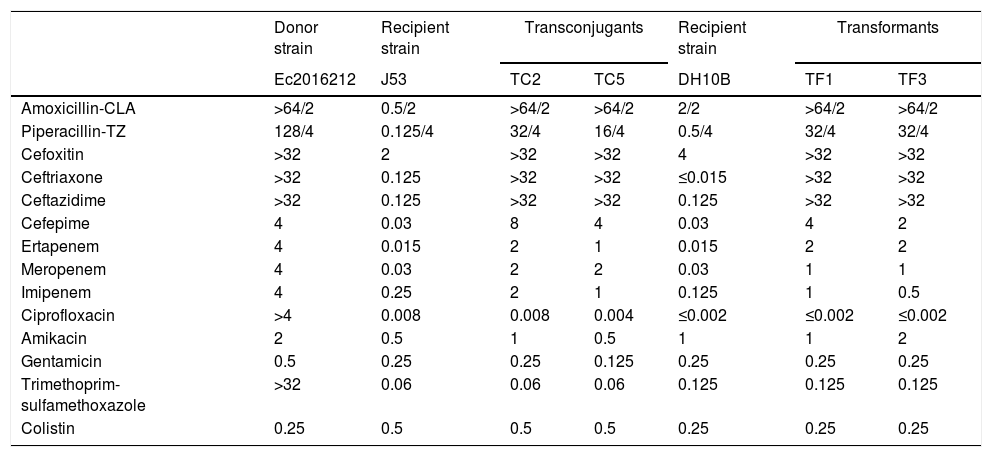
ResultsBoth E. cloacae complex isolates were resistant or showed reduced susceptibility to β-lactam agents (Table 1). The βCarba test was positive and a positive synergy meropenem/dipicolinic acid was observed. PCR detection of ESBL, AmpC and carbapenemase genes indicated presence of only blaNDM.
MICs (mg/L) for the NDM-carrying E. cloacae complex clinical isolate Ec2016212, E. coli J53, E. coli DH10B, transconjugants and transformants.
| Donor strain | Recipient strain | Transconjugants | Recipient strain | Transformants | |||
|---|---|---|---|---|---|---|---|
| Ec2016212 | J53 | TC2 | TC5 | DH10B | TF1 | TF3 | |
| Amoxicillin-CLA | >64/2 | 0.5/2 | >64/2 | >64/2 | 2/2 | >64/2 | >64/2 |
| Piperacillin-TZ | 128/4 | 0.125/4 | 32/4 | 16/4 | 0.5/4 | 32/4 | 32/4 |
| Cefoxitin | >32 | 2 | >32 | >32 | 4 | >32 | >32 |
| Ceftriaxone | >32 | 0.125 | >32 | >32 | ≤0.015 | >32 | >32 |
| Ceftazidime | >32 | 0.125 | >32 | >32 | 0.125 | >32 | >32 |
| Cefepime | 4 | 0.03 | 8 | 4 | 0.03 | 4 | 2 |
| Ertapenem | 4 | 0.015 | 2 | 1 | 0.015 | 2 | 2 |
| Meropenem | 4 | 0.03 | 2 | 2 | 0.03 | 1 | 1 |
| Imipenem | 4 | 0.25 | 2 | 1 | 0.125 | 1 | 0.5 |
| Ciprofloxacin | >4 | 0.008 | 0.008 | 0.004 | ≤0.002 | ≤0.002 | ≤0.002 |
| Amikacin | 2 | 0.5 | 1 | 0.5 | 1 | 1 | 2 |
| Gentamicin | 0.5 | 0.25 | 0.25 | 0.125 | 0.25 | 0.25 | 0.25 |
| Trimethoprim-sulfamethoxazole | >32 | 0.06 | 0.06 | 0.06 | 0.125 | 0.125 | 0.125 |
| Colistin | 0.25 | 0.5 | 0.5 | 0.5 | 0.25 | 0.25 | 0.25 |
Amoxicillin-CLA: Amoxicillin-clavulanate; Piperacillin-TZ: Piperacillin-tazobactam.
PFGE-XbaI showed identical patterns in both E. cloacae complex isolates.
The assembled data from Ec2016212 provided an allelic profile that was assigned to a new MLST type, ST932. The antimicrobial associated resistance genes identified were blaNDM-1; blaCMH-3; aph(3′)-Ia, aadA16, strA and strB; aac(6′)Ib-cr; mph(A), tet(A), sul1 and dfrA27. Plasmid Finder identified IncX3 and IncR replicons. S1-PFGE analysis revealed three plasmids, one of ∼50-kb and two small plasmids <7kb. The transformants and transconjugants only contained the 50-kb plasmid, showing signal with NDM and IncX3 probes, suggesting that blaNDM-1 was in that plasmid.
Conjugation assays confirmed that blaNDM-1 could be transferred between E. cloacae complex and E. coli with a transfer frequency of 7×10−7 per donor. All transconjugants and electroporants showed similar phenotype regarding β-lactams and the presence of the blaNDM gene was confirmed by PCR (Table 1).
blaCMH-3 could not be transferred between E. cloacae complex and E. coli and PCR assays did not detect blaCMH-3 in any transconjugant or electroporant.
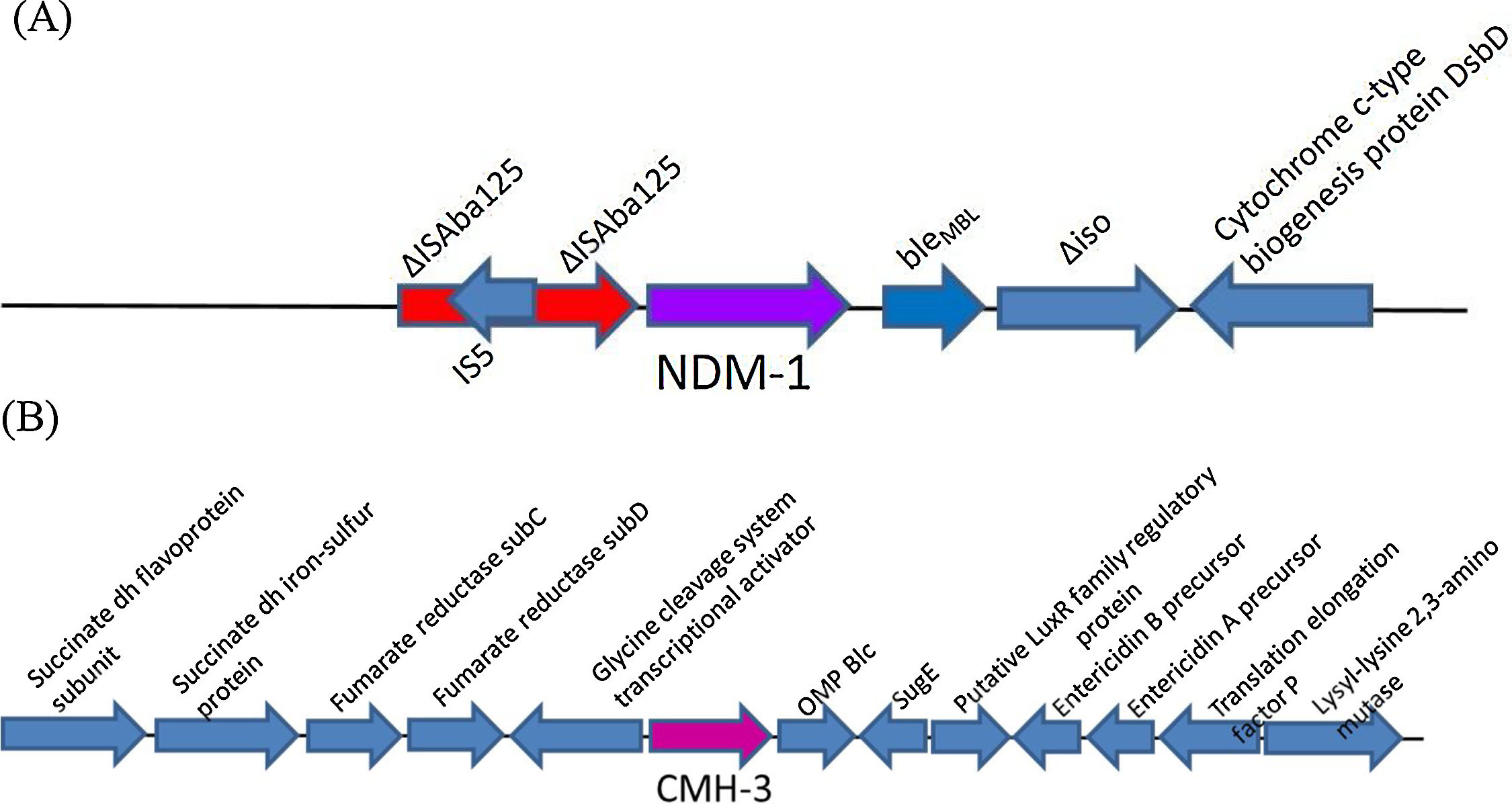
blaNDM gene was surrounded by a truncated Tn125 (ΔTn125) structure (Fig. 1). The analysis revealed that upstream of blaNDM-1 appeared the insertion sequence ISAba125 disrupted by the IS5 element. Downstream the blaNDM-1 were located bleMBL, trpF and dsbC genes and further downstream was an IS26 element. The region flanking blaNDM-1 was like that described for other blaNDM genes.14
The blaCMH-3 genetic background is shown in Fig. 1. A protein BLAST analysis performed between the enzyme detected in Ec2016212 and the known variants of CMH showed 100% identity to CMH-3, 99% to CMH-1 and 98% to CMH-2.
DiscussionThe present study investigated the molecular characterization of an NDM-1 producing E. cloacae complex isolate obtained from a patient transferred from Ukraine to Spain. The strain belonged to a new clone ST932. In Spain, NDM is not the predominant carbapenemase in Enterobacter spp.7,8 A recent work showed that blaNDM producers Enterobacter spp. were predominant in the Balkans, India and Vietnam.4 In accordance with this, our isolate recovered after the transfer from another hospital located in one of these geographic regions suggest that the patient was colonized during his hospitalization in Ukraine. NDM-1 in Enterobacterales is usually located in a plasmid genetic platform and it has been reported that blaNDM-1 is carried by various types of plasmids such us IncF, IncA/C, IncL/M and IncX3.15 The analysis of Ec2016212 showed that blaNDM-1 was carried by an IncX3 plasmid, which was readily transferrable to recipient E. coli J53 by conjugation suggesting the possibility of dissemination. Moreover, a recent work has confirmed the in vivo horizontal transfer of blaNDM carrying plasmids in Enterobacterales.15
NDM-1 harbouring strains from different geographic area have shown similar genetic environments.14,15 In Ec016212 the blaNDM-1 genetic surrounding compared with GenBank sequences revealed that the immediate NDM genetic environment of the IncX3 plasmid was highly similar to others IncX3.14 The presence of this highly conserved region detected in other NDM harbouring bacteria also supports the horizontal transmission.
The other interesting feature of the strain was the presence of CMH-3, which belongs to CMH enzymes, a new group of AmpC-type β-lactamases. To date, three variants have been described (CMH-1, 2 and 3). In 2015, CMH-1 was reported in an E. cloacae isolate from Taiwan, it was successfully conjugated and showed a 99% similarity to a chromosome-borne AmpC in E. cloacae ATCC13047.3 Unfortunately, the biochemical characterization was not studied. In 2017 the variant CMH-2 was described as a plasmid-encoded enzyme in clinical isolates of Klebsiella pneumoniae collected in India.2 CMH-2 showed a 98.6% identity to CMH-1 and provided non inducible resistance to cefotaxime, ceftriaxone and ceftazidime. Transformants and transconjugants were obtained confirming the presence of CMH-2 in a plasmid (IncK replicon).
Regarding CMH-3, the only available information is the nucleotide sequence submitted in 2016 to Genbank (accession number KX192165) from USA without information provided about the genetic location. In Ec2016212 we were unable to determine definitively whether the gene was located on a plasmid or encoded on a chromosome, but the analysis of Illumina contigs that included the enzyme in showed that the enzyme was probably chromosome encoded. No transformants or transconjugants could be obtained in repeated experiments supporting the non-transferability of blaCMH-3. To our knowledge this is the first report of the presence of a CMH-type enzyme in a clinical isolate collected in Europe. In Ec2016212, apart from CMH-3, we detected another chromosomal AmpC showing <80% of homology to CMH-3, but 99% to AmpC detected in the chromosome of other E. cloacae complex isolates. Further studies focused on how the presence of both AmpC enzymes in terms of regulation and/or expression could influence the susceptibility would be challenging. Plasmid-mediated AmpC originated from chromosome of E. cloacae complex have been described.1 In our isolate, knowing that variants CMH-1 and CMH-2 have been found in plasmids it is not unreasonable hypothesize that CMH-3 (if chromosomal location) could be transferred to a plasmid and disseminated in the community.
In summary, the characterization of this E. cloacae complex isolate harbouring NDM in an IncX3 plasmid alerts about the spreading of the enzyme in our geographic area. The detection of CMH-3 in a more than likely chromosomal location and its potential to relocation in a plasmid must be considered. According to this, further assays including kinetics and binding affinities would be interesting to define the role of CMH-3 in the susceptibility to beta-lactams.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethical approvalNot required.
Competing interestsNone declared.
This work was supported by Plan Nacional de I+D+i 13-2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Ciencia, Innovación y Universidades, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0001) cofinanced by European Development Regional Fund “A way to achieve Europe”, Operative program Intelligent Growth 2014–2020.