SARS-CoV-2variants of concern (VOC) have been described in the UK (B.1.1.7), South Africa (B.1.351) and Brazil (P.1). Among them, the most scarce information has been obtained from the P.1 variant and more data on its global presence and about its spreading dynamics are needed.
MethodsWhole genome sequencing was performed prospectively on travellers arriving from Brazil and on a random selection of SARS-CoV-2 positive cases from our population.
ResultsIn this study we report the first SARS-CoV-2 P.1 and P.2 variants exported from Brazil to Spain. The case infected with the P.1 variant, who had only stayed in Rio de Janeiro, required hospitalisation. The two P.2 cases remained asymptomatic. A wider distribution for P.1 variant beyond the Brazilian Amazonia should be considered. The exportation of the P.2 variant, carrying the E484K mutation, deserves attention. One month after the first description of P.1 and P.2 importations from Brazil to Madrid, these variants were identified circulating in the community, in cases without a travel history, and involved in household transmissions
ConclusionWhole genome sequencing of SARS-CoV-2 positive travellers arriving from Brazil allowed us to identify the first importations of P.1 and P.2 variants to Spain and their early community transmission.
Se han descrito «variantes de preocupación» (VOC) de SARS-CoV-2 en el Reino Unido (B.1.1.7), Sudáfrica (B.1.351) y Brasil (P.1). Entre ellas, se dispone de información más escasa para la variante P.1 y se necesitan más datos sobre su presencia global y sobre su dinámica de expansión.
MétodosSe realizó secuenciación del genoma completo de forma prospectiva de SARS-CoV-2 en viajeros procedentes de Brasil y en una selección aleatoria de casos positivos de SARS-CoV-2 de nuestra población.
ResultadosEn este estudio reportamos las primeras variantes de SARS-CoV-2 P.1 y P.2 exportadas desde Brasil a España. El caso infectado por la variante P.1, que solo había permanecido en Río de Janeiro, requirió hospitalización. Los 2 casos de la variante P.2 permanecieron asintomáticos. Se debe considerar una distribución más amplia para la variante P.1 más allá de la Amazonía brasileña. La exportación de la variante P.2, que porta la mutación E484K, merece asimismo atención adicional. Un mes después de la primera descripción de las importaciones de P.1 y P.2 de Brasil a Madrid, se identificaron estas variantes circulando en la comunidad, en casos sin antecedentes de viaje, e implicadas en transmisiones domiciliarias.
ConclusiónLa secuenciación de genoma completo de viajeros positivos para SARS-CoV-2 procedentes de Brasil nos permitió identificar las primeras importaciones de variantes P.1 y P.2 a España y su transmisión comunitaria precoz.
SARS-CoV-2 variants of concern (VOC) have been described in the UK (B.1.1.7), South Africa (B.1.351), and Brazil (P.1).1 They harbour a constellation of mutations with potential significance, some mapping in the receptor binding domain of the spike protein, possibly involved in immune evasion or increase of the affinity for ACE2 receptor.2,3
The fact that different SARS-CoV-2 lineages share relevant mutations suggests a convergent evolution towards an advantageous phenotype. A more efficient transmission rate and an increased mortality4 have been found for B.1.1.7, the most extensively studied VOC, currently being the major lineage in the UK,5 and present in 83 countries.1
Not so much information is available for the P.1 variant.6 Sequencing efforts for P.1 have been less intensive in Brazil than for B.1.1.7 variant in the UK. In the moment of writing this article it had only been detected in 940 cases from 26 countries, with barely two exportation events reported, to Japan and Italy.7,8 Therefore, more data on the global presence of the P.1 variant and about its spreading dynamics are needed.
We present the analysis of three travellers who flew from Brazil to Madrid separately on January 29 and 31, 2021. All were asymptomatic upon arrival and were diagnosed at the airport (antigenic test) within a screening program. They were transferred to our hospital and positive RT-PCR (TaqPath COVID-19, ThermoFisher) confirmed the initial diagnosis.
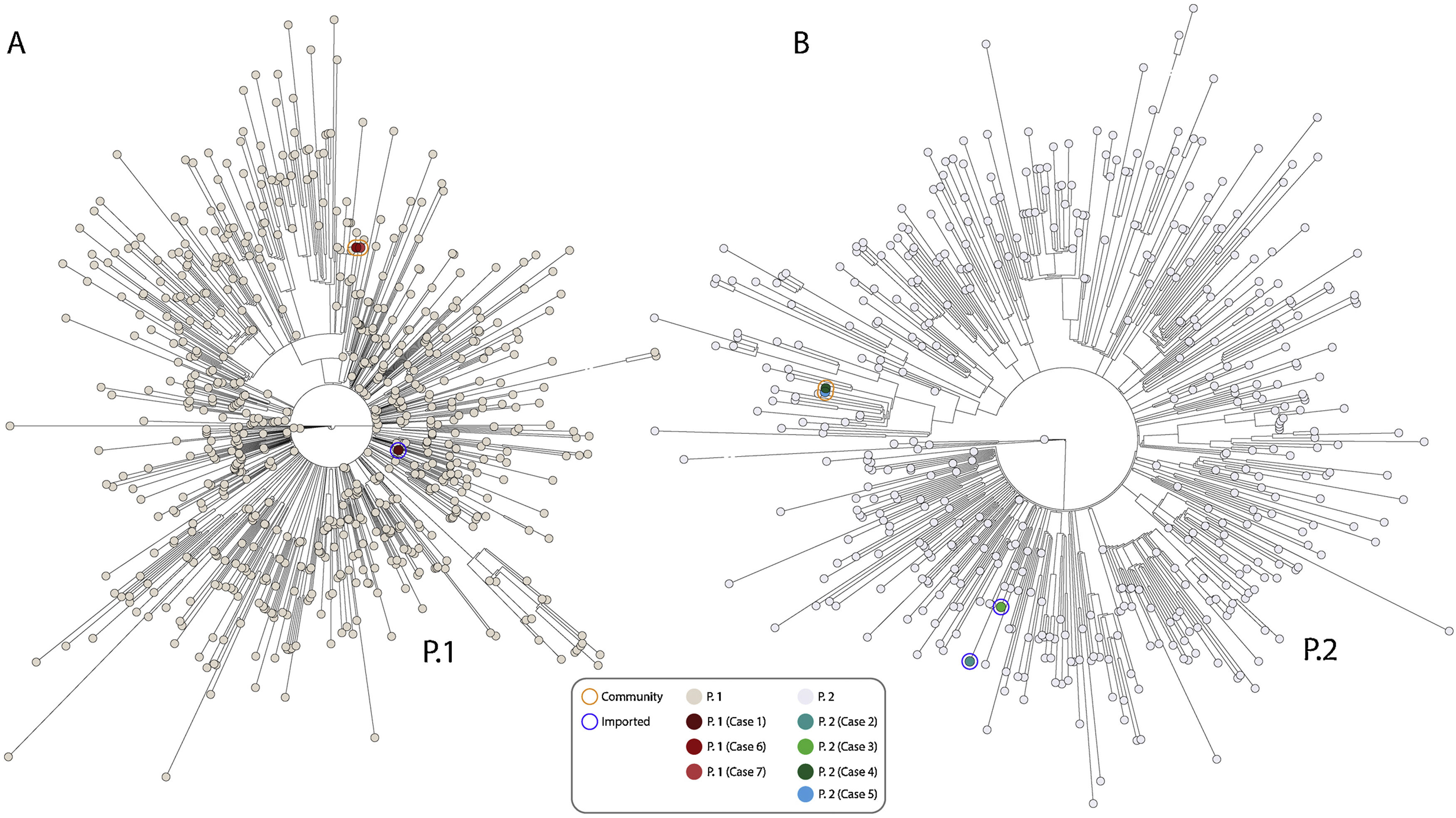
Due to the alarm of the potential import of the P.1 VOC from Brazil, urgent sequencing was activated. Whole genome sequencing was performed with the Artic_nCov-2019_V3 panel of primers. Libraries were prepared using the Nextera Flex DNA Library Preparation Kit (Illumina lnc, California, USA) and sequenced in a NextSeq. Four working days after the diagnosis, we were able to characterise by WGS the variants involved. It allowed us to determine that one of the travellers (Case 1) carried the P.1 variant (Fig. 1A), after confirming that it harboured all 17 P.1 marker-SNPs.
Phylogenetic relationship of SARS-CoV-2 based on the alignment of all P.1 (panel A) and P.2 sequences (panel B) from Brazil (completed (>29000bp) with high coverage (<1% Ns)) deposited in GISAID up to March 15, 2021 (714 and 477 samples respectively). Maximum likelihood unrooted tree displaying the genetic distance between all samples collected and the new presented in this study, colored by their lineage, was performed by a multi-alignment with mafft and IQ-TREE GTR+F+R2 mode with 1000 bootstrap replications. Reference for our sequences in GIDAID are: EPI_ISL_985404 (P.1), EPI_ISL_985408 (P.2), EPI_ISL_985409 (P.2), EPI_ISL_1233413 (P.2), EPI_ISL_1381872 (P.1), EPI_ISL_1477061 (P.1) and EPI_ISL_1647839 (P.2).
Case 1 was a 44-year-old male who had stayed in Rio de Janeiro for 15 days living with another relative who did not develop COVID-19. He had no contact with COVID-19 cases nor other social interactions. He developed symptoms (fever, cough, pain in the left hemithorax) two days after his positive RT-PCR and had to be hospitalised the day after the symptoms began, due to a bilateral pneumonia, requiring supplemental oxygen. He was treated with methylprednisolone and baricitinib (hepatitis ruled out anakinra/tocilizumab), with clinical improvement. For the P.1 case reported in Japan,7 only the father (in his 40s) of the travelling family developed relatively mild symptoms, while the remaining three relatives remained asymptomatic. The other published case (33 years of age), who exported P.1 to Italy, remained asymptomatic.8
The P.1 case reported in this study had only stayed in Rio and the case reported in Italy had not left Sao Paolo. These data indicate that the P.1 variant is probably more widely distributed in Brazil and not only restricted to the Amazonia as initially considered.9
Two of the other positive cases (Cases 2 and 3), were infected with the SARS-CoV-2 P.2 variant; all five P.2 marker SNPs were identified (Fig. 1B).6 Another P.2 sequence had been deposited in GISAID from a case diagnosed on December 27th 2021 in the Canary Islands.
Case 2 was a 44-year-old male who stayed in Rio de Janeiro for 10 days. He remained asymptomatic. Case 3 was a 23-year-old male who travelled to Fortaleza for 15 days to visit his partner. He stayed in a hotel and did not have other social interactions while in Brazil. He travelled back to Madrid via Sao Paolo and spent 6h at the transfer airport. He remained asymptomatic throughout his quarantine. His partner also kept quarantine in Brazil and did not acquire the infection.
The P.1 and P.2 variants have evolved independently from lineage B.1.1.28 in Brazil.6,9 P.2 has not received an equivalent treatment according to global alerts, probably because it does not carry the accumulation of risk mutations found in P.1. However, P.2 carries the E484K substitution, shared by the P.1 and B.1.351 variants, which maps in the RBD (receptor binding domain) of the spike protein. A recent alarm has raised in the UK due to the acquisition of the same E484K mutation in 28 cases infected with the B.1.1.7 variant.1
One month after the identification of the P.1 and P.2 importation events we performed the analysis by WGS of a random selection of 50 SARS-CoV-2 positive cases (10.7%) from the 467 COVID-19 cases with SARS-CoV-2 positive specimens obtained from our population between February 26 and March 4, 2021. The P.2 variant was identified in a 52-year-old Spanish woman (Case 4), without links to Brazil or any history of travelling, indicating community transmission for the P.2 variant. A telephonic epidemiological survey suggested that the potential source might be the Peruvian woman (also without a history of travelling) taking care of Case 4's father, who also had a positive SARS-CoV-2 RT-PCR (specimen not available), as the alert for the Brazilian variants has been recently extended to other countries in Latin America, including Peru (https://cov-lineages.org/global_report_P.1.html). The P.2 variant was also identified in Case 4's father (Case 5; 0 SNPs with respect to Case 4's sequence, indicating household transmission).
Ten days after the identification of the first P.2 community cases, we performed a new weekly WGS analysis on a random selection of 50 SARS-CoV-2 positives cases (13%) from the 385 COVID-19 cases with SARS-CoV-2 positive specimens obtained from our population between March 15 and March 21, 2021. A P.1 variant was identified in a 75-year-old man (Case 6). Both, his 50-year-old daughter (Case 7) and his partner, were also SARS-CoV-2 RT-PCR positive at the time of Case 6's diagnosis. The P.1 variant was also identified from his daughter's specimen (0 SNPs between the P.1 variants from Cases 6 and 7, indicating household transmission), whereas the high Ct values for his partner's specimen did not allow performing WGS. In the telephonic epidemiological survey, they did not refer any history of travelling or contacts with other travellers or foreigners, indicating the community nature of these P.1 variants.
ConclusionsWe report three independent COVID-19 imported cases from Brazil to Spain. To the best of our knowledge, one represents the first imported P.1 SARS-CoV-2 variant to Spain and was associated to a severe disease. The patient had only stayed in Rio de Janeiro, for which reason a wider distribution for the P.1 variant, beyond the Amazonia, should be considered in Brazil. The P.2 variant carrying the E484K concern mutation is also reported here, imported by two other travellers who remained asymptomatic. One month after the first description of P.1 and P.2 importations from Brazil to Madrid, these variants were identified circulating in the community, in cases without a travel history, and involved in household transmissions. Our findings add information to the scarce data available to date on SARS-CoV.2 concern variants emerging in Brazil.
Author's contributionsPedro J. Sola Campoy, Sergio Buenestado-Serrano: Bioinformatic analysis, data analysis, MS revision.
Laura Pérez-Lago: Data analysis, MS revision.
Cristina Rodriguez-Grande: experimental tasks.
Pilar Catalán: diagnostics, PCR, data analysis.
Luis Alcala: databases, data analysis, sample selections.
Cristina Andrés-Zayas: sequencing.
Carmen Losada, Carla Rico-Luna: diagnosis, PCRs.
Patricia Muñoz: resources, network coordination.
Darío García de Viedma: conceptualisation, data analysis, design, MS writing.
FundingThis work was supported by Instituto de Salud Carlos III (Ref COV20/00140: SeqCOVID – Consorcio para la epidemiología genómica de SARS-CoV-2 en España) and by Consejo Superior de Investigaciones Científicas (CSIC) (PTI Salud Global).
Conflict of interestsThe authors declare no conflict of interests.
We are grateful to Dainora Jaloveckas (cienciatraducida.com) for editing and proofreading assistance. We are indebted to Rocío Fernandez del Rey and Maricela Valerio for her valuable help to acquire epidemiological and clinical data.
Adán-Jiménez (Javier), Alcalá (Luis), Aldámiz (Teresa), Alonso (Roberto), Álvarez (Beatriz), Álvarez-Uría (Ana), Arias (Alexi), Arroyo (Luis Antonio), Berenguer (Juan), Bermúdez (Elena), Bouza (Emilio), Buenestado-Serrano (Sergio), Burillo (Almudena), Candela (Ana), Carrillo (Raquel), Catalán (Pilar), Cercenado (Emilia), Cobos (Alejandro), de la Cueva (Víctor Manuel), Díez (Cristina), Egido-Balzategui (Jose), Escribano (Pilar), Estévez (Agustín), Fanciulli (Chiara), Galar (Alicia), García (M a Dolores), García de Viedma (Darío), Gijón (Paloma), González (Adolfo), Guillén (Helmuth), Guinea (Jesús), Haces (Laura Vanessa), Herranz (Marta), Kestler (Martha), López (Juan Carlos), Losada (Carmen Narcisa), Machado (Marina), Marín (Mercedes), Martín (Pablo), Martín-Escolano (Javier), Molero-Salinas (Andrea), Montilla (Pedro), Muñoz (Patricia), Olmedo (María), Otero-Sobrino (Álvaro), Padilla (Belén), Palomo (María), Parras (Francisco), Pérez-Granda (María Jesús), Pérez-Lago (Laura), Pérez (Leire), Pescador (Paula), R Maus (Sandra), Reigadas (Elena), Rico-Luna (Carla Margarita), Rincón (Cristina), Rodríguez (Belén), Rodríguez (Sara), Rodríguez-Grande (Cristina), Rojas (Adriana), Ruiz-Serrano (María Jesús), Sánchez (Carlos), Sánchez (Mar), Serrano (Julia), Sola-Campoy (Pedro J), Tejerina (Francisco), Valerio (Maricela), Veintimilla (M a Cristina), Vesperinas (Lara), Vicente (Teresa), de la Villa (Sofía).