Introduction
Testing for HIV resistance to antiretroviral drugs has been recommended to help the guide of new regimens after failure of treatment 1-3. HIV resistance testing involves genotypic assays, phenotypic assays or both 4. Genotypic assays detect drug resistance mutations in the relevant viral genes (i.e. reverse transcriptase and protease) that are known to confer drug resistance. Interpretation of test results requires knowledge of the mutations or consultation with a specialist in HIV drug resistance in order to select active drugs. Phenotyping assays measures virus's ability to grow in different concentrations of antiretroviral drugs. Drug concentrations that inhibit 50% and 90% of viral replication (i.e. the median inhibitory concentration [IC] IC50 and IC90) are calculated, and the ratio of the IC50 of test and reference viruses is reported as the fold increase in the IC50 (i.e. fold increase). Interpretation of phenotyping test is complicated by the paucity of data regarding the specific resistance level that is associated with drug failure. Again, consultation with a specialist can be useful for interpreting test results.
Resistance testing, both genotyping and phenotyping, has some drawbacks that include the lack of uniform quality assurance for all available assays, relatively high cost and insensitivity for minor viral species 5. Moreover, randomized clinical trials have shown conflicting results when the efficacy of antiretroviral regimes guided by resistance testing was compared with standard of care.
A previous meta-analysis has compared the efficacy of resistance testing with the standard of care to achieve a HIV-1 RNA below the detection limit 6. In the present meta-analysis, we updated the information available, and in addition we assessed the magnitude of HIV1-RNA decrease and the CD4 lymphocyte recovery in the intervention and control groups.
Methods
Eligibility criteria
Studies of HIV infected patients with viral load > 400 copies/mL after at least 12 weeks of any HAART therapy who were randomized to changes in therapy according to results of resistance testing (genotype, phenotype) or therapy changes according to the standard of care. Outcomes evaluated were the proportion of patients with HIV-1 RNA below the detection limit, changes in HIV-1 RNA and, changes in CD4+ cells at the end of follow-up.
Search strategy
We searched (1996-October 2004) MEDLINE and EMBASE using the search strategy (Resistance testing OR Susceptibility testing OR Drug Resistance OR Genotypic Resistance OR Phenotypic Resistance OR Phenotype or Genotype) AND Randomized controlled trial AND HIV. We also searched AIDSLINE and Cochrane Library databases. Abstracts presented at major infectious diseases meetings between 1998 and 2004 were hand searched.
Selection
Each of us independently reviewed each title or abstract, to identify relevant articles. We further independently assessed relevant citations for inclusion using the full publication, or abstracts, if they were never published in full. We measured our agreement on selecting articles for further evaluation and for finally including studies. Disagreement was resolved by consensus. Duplicate or updated publications were identified. We included only the most complete data set in our review. The methodological quality of the included trials was scored according to the validated Jadad 5 point scale 7. The scale consisted of three items describing randomization (0-2 points), masking (0-2 points), and description of dropouts and withdrawals (0-1 points) in the report of randomized-controlled trials. Higher scores indicate better reporting.
Data extracted
Two of us (J.E. and R.F.R.A.) independently selected studies and collected data regarding study quality, baseline HIV-1 RNA, lower limit of detection of HIV-1 RNA, baseline CD4 lymphocyte count, type of resistance testing used, genotype interpretation by experts, time to outcome evaluation, proportion of patients with HIV-1 RNA below the limit of detection at the end of follow-up and differences in CD4+ lymphocytes and HIV-1 RNA between baseline and end of follow-up. Disagreement was resolved by consensus.
Statistical analysis
We calculated the relative risk of achieving a non detectable viral load and its 95% confidence interval for each study using as the numerator the number of patients with no detectable viral load and as denominator the number of patients entered in the study (intention-to-treat-principle). For every trial we calculated the relative risk for the primary outcome. We used I 2 and chi square test to assess if significant heterogeneity was present between the included trials. This statistic tests the null hypothesis that the underlying effect measured by the pooled studies is equivalent. If no significant heterogeneity was present (p values greater than 0.1) we used the fixed effect model to combine the effect sizes of the included studies. In case of finding heterogeneity a random effects model was used to combine the effect sizes 8. We carried out a sensitivity analysis according to the following "a priori" hypotheses: 1) Type of resistance test (genotype vs. phenotype), 2) Genotype test (interpreted by experts vs. not interpreted), 3) Follow-up period (< 24 weeks vs. 24 weeks), 4) Proportion of patients with only a first failure to antiretroviral regimens (≥ 45%, < 45%, or Unknown), and 5) Studies quality assessment according the Jadad scale (< 3 points vs. ≥ 3 points). To compare the effects of resistance testing in the aforementioned subgroups we used the approach described by Altman and Bland 9. Funnel plot review provided a strategy to assess publications bias.
For outcomes that included continuous variables such as HIV-1 RNA reduction and increase in CD4 lymphocyte cell count we used as summary statistic the weighted mean difference. The effect size for each individual study was defined as the difference between the resistance testing group and the standard of care group divided by the pooled standard deviation. Effect sizes were combined to compute a weighted mean effect size.
Results
Trial flow
Figure 1 describes the flowchart diagram. We identified 6 full-length reports in peer-reviewed journals, and two abstracts from conferences proceeding meetings with usable information for the meta-analysis 10-17.
Figure 1. Flowchart diagram.
Study characteristics
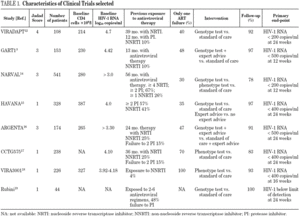
Study characteristics are summarized in table 1. Among the studies, 5 used genotype testing, 2 phenotype testing and 1 both types of resistance testing. The description of standard of care varied among the trials: two studies did not define this term 15,17, three described that standard of care treatment was chosen by clinicians or investigators 11,12,16, and three defined standard of care as an optimum care based on published guidelines, past pharmacological history and virological and immunological parameters 10,13,14.
The length of follow up was equal or less than 24 weeks in all studies. The quality of methods of 3 trials was rated high (≥ 3 points), in the remaining 5 studies allocation was not conceal or was unknown. The funnel plot showed no significant publication bias.
Outcomes
The primary outcome of all studies was the proportion of patients with HIV-1 RNA below the detection limit at the end of follow up. Five trials described the reduction observed in HIV-1 RNA (mean and standard deviation, log10 copies/mL) and three the changes in CD4 lymphocyte count (mean and standard deviation, cells x10 9/L) observed at the end of follow-up (table 2).
Undetectable virus load
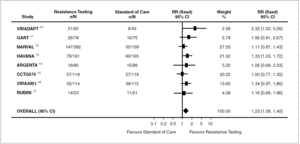
A total of 413 (40.21%) patients in whom therapy was guided by resistance testing achieved a not detectable viral load at end of follow up compared with 258 (32.95%) patients treated according the standard of care (Relative Risk 1.23; 95% CI: 1.09-1.40, p = 0.0009) (fig. 2). Although there were differences among trials in type of resistance test used, rate of virological failure at entry and lower limit of HIV-1 RNA detection, we did not detect significant heterogeneity among trials (p = 0.46; I 2 = 0%). The Absolute Risk Reduction was 8.1% (95% CI; 3%-12%), and accordingly the Number Needed to Treat was 13 (95% CI: 9-25).
Figure 2. Relative risk of achieving a not detectable viral load for all eight trials.
Among our "a priori" hypotheses tested in the subgroup analysis, we found that patients having genotype testing interpreted by experts received the greatest benefit (table 3). The proportion of patients achieving a not detectable viral load at the end of follow-up in patients with therapy guided by genotype resistance testing interpreted by experts compared with the standard of care was 51.6% vs. 29.6, respectively (p = 0.06). Accordingly, the number of patients needed to treat was 5 (95%CI: 3-9).
Studies with intrinsically smallest bias (rated as "high quality") showed the strongest association between resistance testing guided antirretroviral therapy and achieving a not detectable viral load at the end of follow up (table 3).
Subgroup analysis did not show differences between genotype and phenotype resistance testing, neither in virological response according length of follow-up, nor in virological response regarding the number of previous antiretroviral failing regimens.
Decrease in HIV-1 RNA
Five of the eight identified trials, with a total of 1,272 patients, contributed data to this outcome 10,12-14,17. Pooling the results there was a significant (p < 0.000001), although clinically modest difference in favor of resistance testing directed therapy compared with the standard of care (weighted mean difference in HIV-1 RNA reduction: 0.36 log10 copies/mL, 95% CI: from 0.25 to 0.46). Nevertheless, there was a significant heterogeneity among trials (I 2 = 79.7%, p = 0.0006). The observed decrease in HIV-1 RNA (weighted mean difference) varied from 0.57 to 1.5 log10 copies/mL in the resistance testing group, and from 0.39 to 2.0 log10 copies/mL in the standard of care group.
Patients treated according to genotype tests interpreted by experts had a reduction in HIV-1 RNA (weighted mean difference 0.37 log10 copies/mL, 95% CI: from 0.12 to 0.62) greater than those in treated by the standard of care (weighted mean difference 0.24 log10 copies/mL, 95% CI: from 0.01 to 0.48).
CD4 lymphocyte recovery
Three trials, with a total of 823 patients, contributed to this outcome 10,14,17. Pooling the results there was a significant (p < 0.0001) but clinically modest smaller CD4 recovery in the group of patients with therapy directed by resistance testing compared with those who received therapy according the standard of care (weighted mean difference 7.63 x 10 9 cells/L, 95% CI: from 11.03 to 4.22). The observed differences in lymphocyte CD4 counts between the resistance testing and the standard of care groups varied from 12.00 x 10 9 cells/L (95% CI: from 19.97 to 4.03) to 0.00 x 10 9 cells/L (95% CI: from 16.77 to 16.77).
Discussion
Our intention was to examine clinical outcomes in terms of proportion of patients achieving a not detectable HIV-1 RNA, quantitative reduction in HIV-1 viral load, and increase in CD4 lymphocytes, after comparing treatment guided by resistance testing or by standard of care in patients failing antiretroviral therapy. Resistance testing confers a statistically significant benefit for achieving a not detectable viral load compared with the standard of care. However, in our study we found an unexpected high overall number of patients needed to treat (it was necessary to treat 13 patients guided by resistance testing results to obtain one additional patient with a viral load below the detection limit). Moreover, when we estimated the additional reduction in viral load obtained in the resistance testing group compared with the standard of care group, it was only of 0.36 log10 copies/mL. There was not a significant increase in CD4 lymphocyte count between groups. These results have been reproduced in a recently published meta-analysis in every outcome analyzed 18. However, in our study we observed a nearly significant benefit of therapy guided by resistance testing with expert interpretation when compared with standard of care.
Resistance testing has been recommended by several agencies for patients switching therapy due to treatment failures in order to maximize the number of active drugs in the new regimen. Several retrospective studies showed the predictive value of genotypic and phenotypic testing. However, in occasions clinical history alone has been sufficient to predict the failure of subsequent therapy, for example, with abacavir in heavily nucleoside-experienced patients 19 or with nelfinavir in patients with prolonged protease inhibitor experience 20. Zolopa and colleagues 21 found that clinical history alone predicted 40 percent of the treatment response in univariate analysis, genotype alone predicted 67 percent of the treatment response, and adding the clinical history into the model in a multivariate analysis the value was raised to 71%. It has been shown that efficacy of a new antiretroviral regimen in patients with previous virological failure is related with the number of available drugs that still maintain activity 22. Our study did not show the inverse association between exposure to antiretroviral drugs (expressed by percent of patients failing a first antiretroviral regimen) and efficacy of salvage regimens (table 3). Nevertheless, we could not properly test this observation in our meta-analysis due to lack of enough information available from individual studies.
It has been demonstrated that once several regimens have failed, the advantages of pursuing an undetectable viral load become more elusive because of the need to use more complex regimens with inconvenient dosing schedules, greater toxicity, and higher costs 23. In fact, many patients with low grade virological failure (less than 10,000 copies RNA VIH-1/mL), and resistance to the three major drug classes-nucleoside-reverse transcriptase inhibitors (NRTI), protease inhibitors (PI), and non-nucleoside reverse transcriptase inhibitors (NNRTI)- maintain stable or rising CD4-cell counts and remain clinically well. An observational study showed that a complete suppression of HIV viremia is not absolutely necessary for achieving immunologic and clinical stability in patients with multiple antiretroviral regimens' failures 24.
Limitations
Reasons for virological failure are multi-factorial, and emergence of viral resistance is just one of them. Poor penetration of drugs into certain body compartments, insufficient adherence, and variable pharmacokinetics within and between individuals can contribute to virological failures. Moreover, the most common shortcomings of genotypic and phenotypic resistance test include unreliable standardization, variable reproducibility of testing methodology, disorganized or confusing reporting formats for test results, and lack of physician expertise in interpretation.
The major limitation of this meta-analysis was the paucity of randomized clinical trial data, and the short follow up period. The sparseness of the data, and the heterogeneity among trials prevented the assignment of definitive conclusions in terms of potency of antiretroviral therapy guided by resistance testing to reduce HIV-1 viral load and to increase CD4 lymphocyte counts. To avoid selection bias, a systematic and comprehensive search was conducted and two reviewers independently evaluated trials for inclusion. There is a possibility of publication and selection bias in any meta-analysis, however, significant publication bias was excluded according the funnel plot.
Conclusion
Resistance testing is a useful tool to gather knowledge about "in vitro" resistance mechanisms to antiretroviral drugs, and to delineate algorithms to manage patients with virological failure. Our study showed that patients with virological failure received greatest benefit from genotype resistance testing with expert advice when compared with the standard of care. However, it should be taken into account that HIV-1 virological failure is a complex problem in which viral suppression and immunologic benefit are related not only to viral resistance, but also to adherence, drug concentration and the number of active available antiretroviral drugs required to build up a salvage regimen.
Study partly presented at the XI Congreso de la Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC) held in Bilbao (May 16-19, 2004) (Abstract 240).
Correspondence: Javier Ena, MD.
Servicio de Medicina Interna. Hospital Marina Baixa.
Avda. Alcalde En Jaime Botella Mayor, 7. 03570 Villajoyosa. Alicante. España.
E-mail: ena_jav@gva.es
Manuscrito recibido el 18-5-2005; aceptado el 13-9-2005.