A study is presented on the presence of quinolone resistance qnrB1 genes in clinical isolates belonging to the largest series of infections caused by OXA-48-producing Klebsiella pneumoniae in a single-centre outbreak in Spain. Evidence is also provided, according to in vitro results, that there is a possibility of co-transfer of plasmid harbouring blaOXA-48 with an other plasmid harbouring qnrB1 in presence of low antibiotic concentrations of fluoroquinolones, showing the risk of multi-resistance screening.
En este estudio caracterizamos la presencia del gen de resistencia a quinolonas qnrB1 en aislados clínicos pertenecientes a la mayor serie de Klebsiella pneumoniae productora de OXA-48 en un brote de un único hospital en España. Este trabajo ofrece evidencias, mediante ensayos de conjugación in vitro, de que es posible la cotransferencia de plasmidos que albergan blaOXA-48 junto con otros plásmidos que contienen qnrB1 en presencia de bajas concentraciones de fluoroquinolonas, mostrando el riesgo de selección de corresistencias.
Klebsiella pneumoniae is a gram-negative rod of the family Enterobacteriaceae and a common cause of community, nosocomial and opportunistic infections. An emerging association between carbapenems and fluoroquinolone (FQ) resistance is a significant problem in managing such infections. The main mechanisms for transferable carbapenem resistance in this microorganism are due to the emergence of carbapenemases (MBL, KPC and OXA-48-like groups).1 Several low-level plasmid-mediated FQ resistance (PMQR) mechanisms have been described to date in K. pneumoniae: Qnr proteins, the Aac(6′)-Ib-cr enzyme, the plasmid-mediated efflux pumps, QepA and OqxAB.2 Since the increase in MIC values produced by these plasmid determinants is less than the concentrations commonly used in commercial panels, it is difficult to detect this sort of mechanism in routine practice using commercial microdilution panels.
OXA-48 is a carbapenem-hydrolysing oxacillinase that was first described in a clinical isolate of K. pneumoniae and then, OXA-48-producing Enterobacteriaceae have been isolated in several countries in Northern Africa, the Middle East and Europe. It is not easy to ascertain the real prevalence of OXA-48-producing enterobacteria since it is difficult sometimes to detect it routinely in the clinical microbiology laboratory due to low carbapenems MIC values. Bacteria expressing blaOXA-48 also commonly express blaCTX-M-15 and have permeability defects; only then they are resistant to carbapenems, particularly ertapenem (ERT). The largest series of infections caused by OXA-48-producing K. pneumoniae in a single-centre outbreak was recently reported in Spain.3 The predominant clone was assigned sequence type (ST) 405 and harboured blaTEM-1, blaSHV-76, blaCTX-M-15, blaOXA-1 and blaOXA-48 genes. In addition to the multiresistant genetic background, the 78.5% of the isolates of this clone showed a pattern compatible (nalidixic acid susceptible and reduced susceptibility to FQ) with qnr genes. Herein we characterize the additional determinants detected on two strains obtained at the beginning of this outbreak (May 2011).
MethodsK. pneumoniae 471 and 971 were selected and belonged to two different clones (ST405 and a sporadic clone) of this outbreak. Both isolates were recovered from clinical samples: one from urine and the other one from a wound sample.
Whole-cell DNA of these isolates was used as a template for PCR amplification. Screening for PMQR genes (qnrA, qnrB, qnrS, qnrC, qnrD, qnrVC, qepA, oqxAB and aac(6′)-Ib-cr) was performed.4 DNA bands compatible with qnrB, aac(6′)-Ib-cr and oqxAB were identified and confirmed by sequencing (with qnrB identified as qnrB1). Furthermore, the QRDR sequences of the gyrA and parC genes did not identify mutations associated with FQ resistance in either of the two isolates.
Localisation of the qnrB, aac(6′)-Ib-cr, oqxA, oqxB, blaOXA-48 and blaCTX-M-15 genes (plasmidic and/or chromosomal) was determined by (i) Southern-blot using electroporated plasmid extracts obtained with the Kieser method; and by (ii) conjugation (performed 3 times) using Escherichia coli J53 AzR and selection on plates containing 100mg/L sodium azide combined with 0.03, 0.06, 0.125, 0.25 or 0.5mg/L of ciprofloxacin (CIP) and/or 0.25mg/L of ERT.5
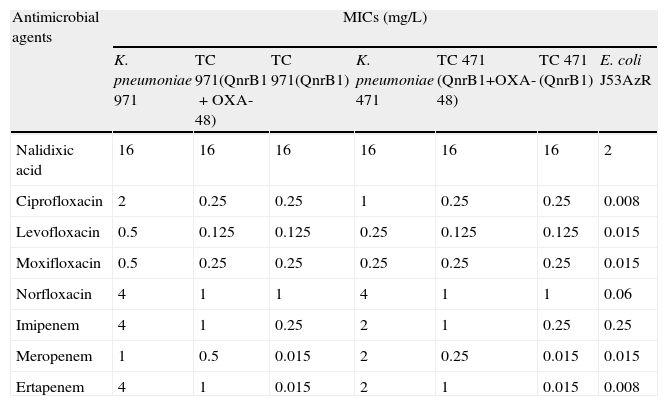
Results and discussionK. pneumoniae 471 and 971 isolates showed reduced susceptibility to FQ (Table 1) but remained susceptible to nalidixic acid.
MICs of FQ and carbapenems for K. pneumoniae 971 and 471 strains and derived transconjugants (TC) in E. coli J53 AzR coding for QnrB1, with or without OXA-48.
| Antimicrobial agents | MICs (mg/L) | ||||||
| K. pneumoniae 971 | TC 971(QnrB1+OXA-48) | TC 971(QnrB1) | K. pneumoniae 471 | TC 471 (QnrB1+OXA-48) | TC 471 (QnrB1) | E. coli J53AzR | |
| Nalidixic acid | 16 | 16 | 16 | 16 | 16 | 16 | 2 |
| Ciprofloxacin | 2 | 0.25 | 0.25 | 1 | 0.25 | 0.25 | 0.008 |
| Levofloxacin | 0.5 | 0.125 | 0.125 | 0.25 | 0.125 | 0.125 | 0.015 |
| Moxifloxacin | 0.5 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.015 |
| Norfloxacin | 4 | 1 | 1 | 4 | 1 | 1 | 0.06 |
| Imipenem | 4 | 1 | 0.25 | 2 | 1 | 0.25 | 0.25 |
| Meropenem | 1 | 0.5 | 0.015 | 2 | 0.25 | 0.015 | 0.015 |
| Ertapenem | 4 | 1 | 0.015 | 2 | 1 | 0.015 | 0.008 |
Transconjugants were obtained in the three selection conditions and three patterns were observed: (1) transconjugants with only reduced susceptibility to FQ and cephalosporins that hybridized with qnrB1, aac(6′)-Ib-cr and blaCTX-M-15, all of which were located on a single 180-kb plasmid (selected with FQ); (2) transconjugants with only reduced susceptibility to carbapenems that hybridized with blaOXA-48 located into a 70-kb plasmid (selected with ERT); and (3) transconjugants with reduced susceptibility to carbapenems, cephalosporins and FQ carrying the two 180- and 70-kb plasmids (selected with CIP plus ERT or only CIP) (Table 1). Sixty percent of the transconjugants selected using only FQs contained also blaOXA-48 gene, independently of concentration of CIP used; besides the frequency of conjugation was higher (10−4–10−5) at low CIP concentrations (0.03–0.125mg/L) when compared to higher CIP concentrations (10−6–10−7 for 0.25–0.5mg/L). oqxA and oqxB genes showed a chromosomal location. Non-typeable incompatibility groups were associated with blaOXA-48 plasmids and a positive result was obtained with a PCR for phage replication protein P (RepP).6blaoxa-48 was associated with Tn1999.2 transposon (JN714122) and qnrB1 was located near to ISCR1.2,7
The blaOXA-48 gene has mainly been described in K. pneumoniae and, as far as we are aware, this is the first time that blaOXA-48 has been associated with the PMQR qnrB1 gene. This data indicate that both plasmids were conjugative. Plasmid size, non-typeability with PCR-based replicon typing and a positive RepP result are features which suggest a relationship with the previously characterized IncL/M epidemic plasmid encoding OXA-48. Here we add evidence, by in vitro assay, that it is possible for the co-transfer of plasmid harbouring blaOXA-48 with another plasmid harbouring qnrB1 in presence of low concentrations of FQ. We have only tested FQ, but further experiments with other antibiotics could also show the risk of selecting for co-resistances. It should be noted that qnrB1, blaOXA-48, aac(6′)-Ib-cr and blaCTX-M-15 were associated in two different plasmids, which together conferred resistance to FQ, β-lactams (including carbapenems) and aminoglycosides.
Conflict of interestThe authors declare no conflict of interest.