Acute respiratory infections are the second cause of morbidity and mortality in children and adults worldwide, being viruses, bacteria and fungi involved in their aetiology. The rapid diagnosis allows for a better clinical management of the patient, for adopting public health measures and for controlling possible outbreaks. The main etiologic agents can be diagnosed within the first hours after the onset of symptoms with antigen detection techniques, primarily immunochromatography. Results are obtained in 15–30min, with 70–90% sensitivity and >95% specificity for the diagnosis of Streptococcus pneumoniae and Legionella pneumophila serogroup O1 infections from urine, Streptococcus pyogenes from throat swabs and respiratory syncytial virus from nasopharyngeal aspirates. Worse results are obtained for influenza viruses and Pneumocystis jirovecii with these techniques; however, other easy-to-perform molecular techniques are available for the rapid diagnosis of these microorganisms. In general, these techniques should not be used for monitoring the outcome or response to treatment.
Las infecciones respiratorias agudas son la segunda causa de morbimortalidad tanto en niños como adultos a nivel mundial en cuya etiología se implican virus, bacterias y hongos. Su diagnóstico rápido permite un mejor manejo clínico del paciente, adoptar medidas de salud pública y controlar posibles brotes. Los principales microorganismos responsables pueden diagnosticarse en las primeras horas tras el inicio del cuadro con técnicas de detección de antígeno, fundamentalmente inmunocromatografícas. Se obtienen resultados en 15–30 min, con una sensibilidad del 70–90% y especificidad superior al 95% para el diagnóstico de infecciones por Streptococcus pneumoniae y Legionella pneumophila serogrupo O1 a partir de orina, Streptococcus pyogenes en exudados faríngeos y virus respiratorio sincitial en aspirados nasofaríngeos. En infecciones por los virus de la gripe y por Pneumocystis jirovecii, los resultados con estas técnicas son peores; no obstante, existen técnicas moleculares de fácil ejecución para el diagnóstico rápido de estos microorganismos. En general, estas técnicas no deben utilizarse para control evolutivo ni para valorar respuesta al tratamiento.
Respiratory tract infections are one of the primary causes of morbidity and mortality and one of the principle reasons for medical consultation all over the world. As in other infectious processes, swift accurate diagnosis is associated with more targeted and effective treatment, lower transmission of the disease and, often, a reduction in its duration.
The aim of this study is to provide a review of rapid diagnostic tests (RDT) in some of the most common respiratory infections (RI). Chronic evolution RIs, such as tuberculosis or those which affect patients in specific situations (e.g. pneumonia associated with respiratory support), have been excluded as they are addressed in other chapters.
RDTs have been considered as those whose result can be given in under 7h (a standard work shift) in comparison with conventional bacterial or viral culture techniques (18–24h and 48h or more, respectively). Even though serological antibody detection techniques meet this temporal definition, they have not been included given that demonstrating seroconversion (appearance of antibodies in the serum) or seroreinforcement (increase in the initial titre of antibodies by a factor of 4) may take weeks or months to come about, with high levels of variability between individuals.
Given the simplicity and swiftness of their implementation and with which results can be obtained and interpreted, some of these RDTs may also be used as “point-of care tests” (POCT) with the subsequent benefits for the patient, who may receive the diagnosis and, depending on the result, the treatment in one single consultation.
In general, the use of RDTs in the diagnosis of RIs can help to:
- •
Reduce the use of antibiotics, given that many RIs are of a viral aetiology.
- •
Ensure the use of suitable anti-viral therapy in specific cases.
- •
Minimise the use of unnecessary diagnostic tests.
- •
Reduce the length of hospital stays.
- •
Permit the swift implementation of isolation measures to limit nosocomial infection, whenever necessary.
Moreover, in RIs of bacterial origin, on detecting the antigen of the target pathogen or its nucleic acids, RDTs are affected to a much lesser extent than culture in the diagnosis in the event that antibiotic treatment has already commenced.
Rapid diagnostic tests for group A Streptococcus (Streptococcus pyogenes–S. pyogenes–)The RDTs for group A Streptococcus (GAS) are fundamentally aimed at determining the bacterial nature of the pharyngitis. Pharyngitis is the most common infection caused by GASs, and may be accompanied by suppurative sequelae (e.g. peritonsillar abscesses) or non-suppurative sequelae (e.g. rheumatic fever and acute glomerulonephritis), although nowadays these complications are rare in the majority of developed countries. Less frequently, they may also lead to serious infections, such as necrotising fasciitis, and other infections such as pneumonia, endocarditis or meningitis.
The majority of paediatric pharyngitis, particularly in children under 3 years of age, are caused by viruses whose symptomatology is very similar to streptococcal infections. It is estimated that only between 20% and 30% of cases of pharyngitis in children, and 10% of those in adults are due to GAS1 and thus a very low percentage of patients will benefit from treatment with antibiotics. This explains why RDTs for the diagnosis of this minor illness are among the most regularly used and are under continual assessment, since they help to prevent the inappropriate use of antibiotics.
The gold-standard test for the diagnosis of GAS-induced pharyngitis continues to be bacterial culture in blood agar, either direct, after enrichment, or with selected plates incubated for between 24 and 48h.
RDTs for the detection of the GAS antigen in pharyngeal swabs appeared at the beginning of the 1980s. Since then, there have been a number of generations of RDTs which have employed different methodologies. Initial techniques employed latex agglutination, followed by the ELISA and lateral flow tests and colorimetric immunochromatographic tests. Recently, molecular tests, such as DNA probes, PCR and in situ fluorescent hybridisation2 have been marketed.
Immunochromatographic tests (ICT) are currently among the most frequently used RDTs owing to their ease of use and rapid results (15min). There are a great many of these tests on the market, but they all detect the C carbohydrate in the GAS cell wall by means of specific monoclonal or polyclonal antibodies. In the majority of these tests, the carbohydrate must be extracted from the cell wall beforehand in an acid medium capable of dissolving them, for their subsequent detection on the immunochromatographic strip.
The optimum performance for these tests is attained in populations previously screened on the basis of certain pharyngeal infection clinical criteria.3 Compared with culture as a reference method, the meta-analyses and studies performed for GAS RDTs in the pharynx give a mean sensitivity of 85% (varying between 70% and 90%) and a specificity of around 95%.1,3–5 Generally speaking, sensitivity varies greatly between studies, but not specificity, which is usually very high. The relatively low sensitivity means that the majority of authors continue to recommend taking a culture in case the test is negative, in order to detect a greater number of cases.6
The great advantage of ICTs is that they can be performed in the presence of the patient, which, along with the high specificity, means that after a positive result no confirmatory culture is required, and specific antibody treatments can be recommended on the spot. Except under special circumstances, performing an RDT or control culture at the end of this treatment is not recommended.
Despite the advantages offered by RDTs, it should be remembered that the culture and isolation of the GAS will facilitate the subsequent performance of other tests, such as the antibiotic sensitivity test or the genotypic characterisation of the isolations. It should also be remembered that, even though its role is subject to debate, in a low percentage of bacterial pharyngitis, beta-haemolytic streptococci from other groups (above all from groups C and G) are isolated which cannot be detected by means of GAS RDTs since the composition of the carbohydrates in their cell wall is different.7
There are also commercial molecular RDTs for the diagnosis of GAS-induced pharyngeal infection through hybridisation and real-time PCR. Although their sensitivity and specificity are as high, or higher, than the culture method, and their limit of detection exceeds that of antigen detection techniques,8 they are still techniques which take between one and two hours to perform, with the need for qualified staff and specific molecular biology equipment. All of this, along with the simplicity and sound diagnostic performance of RDTs for the ICT antigen, has meant that there are currently very few clinical laboratories routinely using molecular techniques such as RDTs for GAS-induced pharyngitis.
For invasive infections caused by GAS, the performance of RDTs has also been compared to culture, with the sensitivity of ICT antigen detection tests being similar to PCR and greater than culture.8 Despite these tests not being recommended in the presence of antibiotic treatment, RDTs may be positive in these samples with negative culture, since they take longer to become negative.
Rapid diagnostic tests for infection by Legionella pneumophila (L. pneumophila)Legionella is a group of bacteria which are ubiquitous in aquatic habitats, comprising more than 55 species and 70 serogroups. Exposure to aerosols containing Legionella can give rise to different clinical manifestations, from Legionnaires’ disease (severe community-acquired pneumonia), to Pontiac fever (self-limited febrile syndrome), or even asymptomatic infection. Of all of these, pneumonia is the most important, owing to its frequency (accounting for around 2–6% of all community-acquired pneumonias (CAP)),9–11 the severity of the illness and the need for specific treatment with macrolide or fluoroquinolone antibiotics. Although it is thought that any species of Legionella can cause illness in humans, and that more than half of the serogroups have been isolated in clinical samples, the vast majority of Legionella-induced pneumonias are caused by L. pneumophila serogroup O1.11,12
The difficulty and slowness of growing Legionella in specific culture media and the time required for seroconversion in the serological diagnosis have meant that the majority of diagnostic tests regularly used in diagnosis of pneumonia caused by L. pneumophila are RDTs. These can be distinguished between those using antibodies and those using nucleic acid amplification tests, essentially PCRs.
The first RDT for legionellosis was direct immunofluorescence (DIF) on respiratory samples using specific serogroup antibodies marked with fluorescein. DIF, which can be considered an RDT since it can be performed in around an hour, has a relatively low sensitivity (60%)13 owing to which it was displaced by the Legionella in urine antigen detection test.
The Legionella antigen in urine can be detected from a few days after the onset of symptoms until months after the infection has been cured, although in the majority of cases it becomes negative between one and two months after the onset of the disease. There are different methods for detecting the Legionella antigen in urine: enzyme immunoassay on 96-well plates (EIA), enzyme-linked immunosorbent assay (ELISA) and the most rapid, ICTs, in either card or strip format, also known as the lateral flow test. These ICTs and ELISA demonstrate sensitivities of around 70–90% and specificities of 95–100%, depending on the study.11,14,15 The majority of commercial tests only detect the L. pneumophila serogroup O1 urinary antigen, although cross-reactions with other serogroups may occur. This implies that pneumonias caused by other species of Legionella (Legionella micdadei, Legionella bozemanii, etc.) or by other serogroups, although much less frequent, will not normally be diagnosed using these tests. There are also other tests which detect more serogroups and species, but their sensitivity and specificity characteristics are lower than for L. pneumophila serogroup O1.
In tests for detecting the Legionella urinary antigen, concentrating the antigen by filtering enhances their sensitivity. On the other hand, boiling the urine can enhance specificity, since it reduces non-specific interactions. Nonetheless, these improvements in rapid tests increase the time and workload until the result is obtained. ICT tests for detecting the L. pneumophila serogroup O1 urinary antigen are considered POCTs since the qualitative results are obtained in minutes, without the need for specialised personnel to take the samples or perform the tests. A fluorescent ICT test has recently emerged which provides greater sensitivity in detection, while eliminating the possible bias attributable to the visual reading of the result.16 The lower specificity observed, particularly in concentrated urine, improves when repeating positive tests after boiling the urine. EIAs are somewhat slower, more quantitative and, in general, used in clinical laboratories which analyse a large number of samples.
In any case, and despite its poorer cost effectiveness, it is important to perform the culture and attempt to isolate the microorganism for subsequent epidemiological and microbiological studies.
Marketed nucleic acid amplification RDTs include isothermal amplification, conventional PCR and real-time PCR on respiratory samples from the lower respiratory tract (bronchoalveolar sputum, aspirate or lavage, etc.), all with high levels of sensitivity and specificity. Other benefits of their use include the rapid response time and their current widespread use in Clinical Microbiology laboratories. Unlike culture, seroconversion and the detection of the urinary antigen, a positive PCR is still not considered as a basis for confirming Legionella infection and must be interpreted with caution.
Legionella genes amplified by the majority of molecular techniques are the genes which encode the 5S and 16S ribosomal RNA subunits, the 16S–23S spacer region and the macrophage inhibitory protein (MIP). With these genes and suitable probes it is also possible to detect and distinguish between L. pneumophila and other species of Legionella.15,17 Nucleic acid amplification tests have shown greater sensitivity (>95%) than urinary antigen tests in the detection of pneumonia caused by Legionella, as well as very high specificities (>95%).15,17
Rapid diagnostic tests for respiratory infection caused by Streptococcus pneumoniae (S. pneumoniae)S. pneumoniae is undoubtedly the pathogen which most frequently causes CAP, particularly its serious forms, when there is bacteraemia and the patient's life is at risk. Unlike Legionella, the presence of S. pneumoniae (pneumococcus) in a respiratory sample is not always indicative of infection, since it is also a known coloniser of the upper respiratory tract, which complicates the interpretation of its diagnosis.
The detection of the pneumococcus antigen in urine is one of the available RDTs. These tests detect the presence in urine of C-polysaccharide in the pneumococcus cell wall, an antigen common to the almost 100 different serotypes of pneumococcus described to date and to other related species, such as Streptococcus mitis and Streptococcus oralis.18 The first tests were described in 1917 and a number of different techniques were tried for its detection (latex agglutination, coagglutination, enzyme immunoassay); however, the results were not promising and they were abandoned.18,19 The emergence of ICT tests with reliable results lead to a reawakening of interest in its detection.20
The sensitivity of the urinary antigen for the diagnosis of pneumococcal pneumonia by means of ICT varies greatly depending on the study, above all owing to the lack of a gold-standard which unequivocally establishes the aetiology of pneumonia. These sensitivities normally vary between 60 and 75%, and are generally higher in bacteraemic pneumococcal pneumonia studies.21–23 Specificity also varies according to the studies, but in general, the results are higher and more homogeneous, ranging from 95 to 99%.21–23
A positive result must be interpreted with caution since it may reflect asymptomatic colonisation, particularly in children, where the performance of this test is not recommended as children tend to be nasopharyngeal carriers. Additionally, the test may be positive months after pneumococcal infection.24 However, the detection of the antigen in urine, unlike in culture, is practically unaffected by prior antibiotic treatment.
Recently, other tests which detect in urine the capsular pneumococcal polysaccharide in pneumonia caused by 14 serotypes (1, 3, 4, 5, 6A/C, 6B, 7F/A, 8, 9V, 14, 18, 19A, 19F and 23F) using Luminex technology have been developed, and although the sensitivity seems greater than that of the detection of C-polysaccharide, they are still not commercially available.25 This test showed a correspondence of only 51% with the C-polysaccharide detection test, and would seem to be a more sensitive alternative when further serotypes are incorporated into the detection.
Other RDTs include the detection of S. pneumoniae DNA through PCR. The high sensitivity of these tests, along with the possibility of colonisation by the pathogen, makes the interpretation of a positive result very difficult. Prominent among the genes used in the detection of S. pneumoniae is the autolysin gene (lytA), the pneumolysin gene (ply) and Spn9802. Attempts have been made to quantify the S. pneumoniae load by real-time PCR in respiratory samples in order to distinguish between colonisation and infection; despite this, no standard criteria have been established for distinguishing between both clinical situations.
In addition to the diagnosis of pneumonia, RDTs for pneumococcus have also been used for detecting the pathogen in sterile samples such as CSF in meningitis26,27 or pleural liquid in parapneumonic effusions.28 The detection of the pneumococcal antigen by ICT in these sterile samples has been shown to be more sensitive than culture, above all as it can still be detected in the presence of antibiotic treatment. On the other hand, pneumococcus detection by means of PCR in sterile samples has been used successfully, given that its presence in these samples, unlike in respiratory samples, constitutes diagnosis of infection.
It should be remembered that despite the advantages of RDTs in the diagnosis of pneumococcal infections, these must supplement rather than replace blood culture and sputum culture, which will make it possible to isolate the causal microorganism, ascertain its antibiotic susceptibility as well as other microbiological characteristics (serotypes, genotypes, etc.) which are of great value in the handling and control of the infection.29
Rapid diagnostic test in viral respiratory infectionsAcute RI of viral aetiology constitutes, as a whole, one of the infectious clinical syndromes which most affects humans. In children, over 50% of CAPs, up to 90% of cases of bronchiolitis and approximately 85% of asthma attacks are associated with infections by respiratory viruses (RV), while in adults RVs have been detected in 20–40% of CAPs, in 50–70% of asthma attacks and between 30–50% of COPD attacks.30,31
Around twenty viruses have been implicated as aetiological agents in these processes. The most important, owing to their clinical epidemiological relevance, are influenza A and B viruses and the respiratory syncytial virus (RSV), and to a lesser extent, adenovirus (AD), rhinovirus (HRV), metapneumovirus and parainfluenza virus 1 to 4 and others, such as enterovirus, coronavirus and bocavirus, etc.
The majority of acute viral RIs are self-limited and progress to recovery with no aetiological treatment whatsoever. Only occasionally (if we exclude certain emerging virus diseases, such as coronavirus (SARS) or new types of influenza) and fundamentally in young or elderly patients or with underlying pathologies susceptible to complications, do we see clinical conditions which, owing to their severity, require special care. Currently, specific anti-viral medicines are used in infection by the influenza virus and in specific clinical situations; for others, such as RSV, HRV or AD, although certain compounds (e.g. ribavirin against RSV and AD and pleconaril for HRV) have shown certain activity, their use has not been sufficiently established.
As a consequence of acute RIs, every year, coinciding with the seasonal epidemics of influenza and RSV, there are excessive demands on the healthcare system, resulting in an increased number of visits and the overloading of the A&E Department, increased consumption of antibiotics and an increase in the performance of analytical and imaging tests.
Based on samples from the respiratory tract (basically nasopharyngeal aspirate or lavage, and to a lesser extent samples taken with nasal and/or pharyngeal brushing in the viral transport medium), there are currently a number of procedures available to us which can help in the rapid aetiological diagnosis of the principal acute viral RIs.
The different techniques can basically be split into two groups: antigen detection tests and nucleic acid amplification tests (NAAT).
Viral antigen detection testsDirect and indirect immunofluorescence (IF) and ICTs are the most commonly used of these tests. Others, such as immunoenzymatic membrane techniques, frequently used in the past, have been pushed into the background.
Using IF it is possible to detect the principal RVs. Nonetheless, conducting IF correctly involves a certain degree of difficulty. The need for a centrifuge to make the extensions, setting and staining them correctly, the need to use a fluorescence microscope to view them, subjectivity in the interpretation of the result (skill of the observer), high dependence on the sample quality (presence of sufficient epithelial cells) and time required to obtain results (between 90 and 120min) means that this technique is being used increasingly sparingly in the initial diagnosis of a viral RI in the majority of clinical laboratories.
The use of classical IF techniques can be improved and facilitated by automating the process and performing a rapid objective reading of the reaction. In this regard, techniques have been developed to detect specific viruses individually32 and others which increase the panel of viruses to be detected, which, apart from RSV and influenza, detects AD, parainfluenza virus 1 to 4, bocavirus, metapneumovirus and bacteria such as pneumococcus. Their possible advantages, usefulness and cost effectiveness have recently been reported, although there are still numerous misgivings in this regard.33
ICT tests which reveal the reaction with colloidal gold are currently the most widely used. Its simplicity (it requiring no special skills) and rapid implementation (results are obtained within 10–20min) and the simple reading of the results are helping to spread their use as procedures to be used rapidly at the patient's bedside or in A&E as POCTs. They are less sensitive than IF, and unlike the latter, which allows a wide range of RVs to be detected, the majority of existing ICT devices have been designed to detect RSV and/or influenza A and B.
With ICT, a great deal of variability is observed in the results, depending on the manufacturer, the sample used (much better sensitivity results are observed with nasopharyngeal aspirate/lavage than with samples taken with a brush), the time of the disease's evolution when the sample is taken (better in the first 24h) and above all, the population in which it is applied (much worse results in the adult population than in children).34–36 In general, the sensitivity of ICT techniques is better when used to detect RSV than in the diagnosis of influenza.
Nucleic acid amplification testsMolecular techniques, basically PCR, are highly sensitive and specific, and have replaced cell culture as a reference method in the diagnosis of infections caused by RVs. They also make it possible to detect non-viable viruses, or those which it has not been possible to isolate in traditional cell cultures. They generally have a response time in excess of 24h and require specific infrastructure and equipment as well as specialised personnel, limiting their use for rapid diagnosis or in emergency situations.37 Nonetheless, in recent years NAATs have been developed in different quick and easy formats, with no loss of efficiency compared to classic molecular procedures. With minimal sample handling and after dispensing the sample into individual devices, the extraction, application and detection of RVs is performed on one single piece of equipment.38–40
As is true of viral antigen detection tests, they have been developed commercially from tests aimed at detecting one specific virus (RSV and/or influenza) evolving into “multiplexes” capable of simultaneously detecting the majority of possible viruses involved in these processes. All this with a result obtention time of between 15 and 90min, with a minimal work load (around 2min), meaning that it is currently possible to use them as POCTs, with results that are much more reliable than those obtained with viral antigen detection tests.31,35,41 In general, these devices make it possible to process a limited number of samples at the same time (between 1 and 8, depending on the platform). Other systems using Luminex technology make it possible to process up to 96 samples and simultaneously detect 22 different pathogens (19 viruses and 3 bacteria), obtaining results in 4–5h; but the need to use three different items of equipment in the processing hinders their use as rapid tests.
Use of rapid tests in the diagnosis of acute respiratory infection of viral aetiologyWith regard to acute viral RIs, we find cases ranging from outpatients requiring a rapid diagnosis of a specific agent (as has occurred with the flu pandemic in pregnant women or the morbidly obese) to hospitalised patients with serious underlying diseases (e.g. onco-haematological conditions), where the most important thing is to rule out any of the possible viruses involved. Apart from the particular situations commented on above, or in outbreaks in sealed-off institutions, there is currently a fairly broad consensus on the non-use of these tests outside the hospital environment.
In emergency care areas, the use of viral antigen detection tests is widespread, particularly for RSV and influenza, but this has not been shown to reduce the prescription of antibiotics or to reduce the number of supplementary diagnostic tests (analytical or imaging). Even in the case of RSV, it can lead to a false sense of security when distributing patients and establishing isolation measures, with results that may at times be counter-productive. It has only been possible to associate the use of these tests in emergencies to lower consumption of antibiotics, but only once patients return to the outpatient setting.42
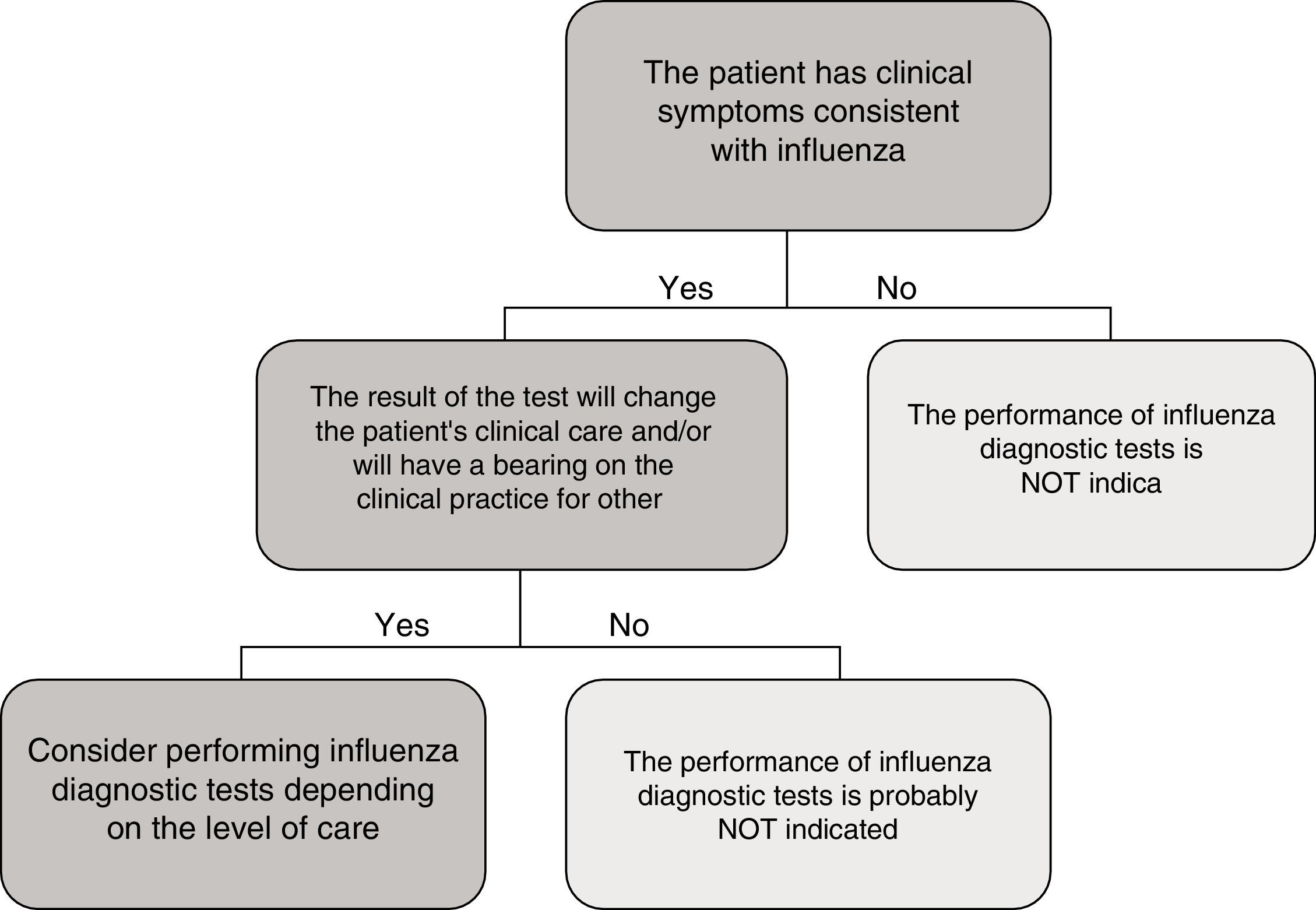
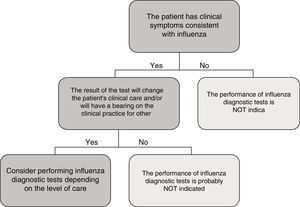
In the specific case of viral antigen detection tests for the influenza virus, they generally demonstrate a highly deficient sensitivity and a negative predictive value, owing to which their use in emergency healthcare, beyond specific high-risk cases and if the patient does not need to be admitted, has been called into question. Fig. 1 shows a proposed algorithm for the use of influenza diagnostic tests during epidemic periods. Generally speaking, if it is considered necessary to rule out influenza virus owing to the patient's clinical situation, in order to start the treatment early or to implement preventive measures for nosocomial outbreaks of influenza, it is better to implement a NAAT directly, just as if the patient required hospital admission.43 Although performing influenza diagnosis tests is recommended in hospitalised patients with compatible clinical symptoms, in these cases, empirical anti-viral treatment must be initiated as soon as possible, without the need to await the result of diagnostic tests.
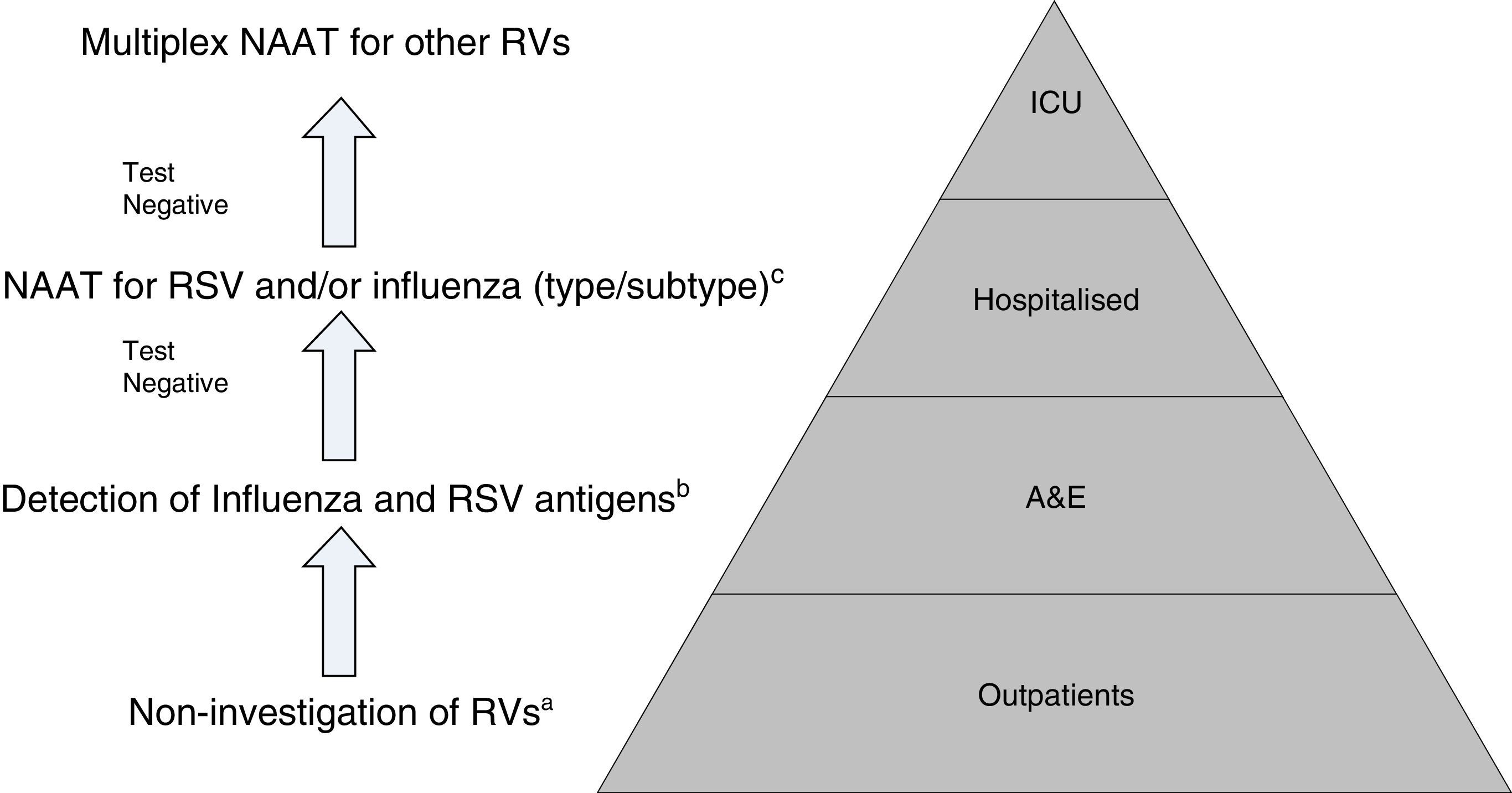
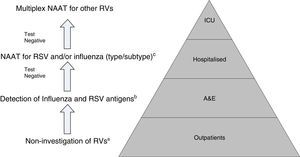
Proposal for a rational work flow and cost-effective algorithm for the investigation of respiratory viruses (RV). a Except for patients included in monitoring programmes (e.g. National Influenza Monitoring Network). b Except in special situations: pregnancy, predisposing underlying disease, etc., in which NAATs will be performed directly. c NAATs for other RV (rhinovirus, metapneumovirus, etc.), may be added in patients with a high risk of serious respiratory illness. Modified by Navarro-Marí et al.45
Owing to their high sensitivity, NAATs are capable of detecting asymptomatic excretion or very low viral loads of dubious clinical significance. Moreover, there are no data to support the use of NAATs for evolutionary control or patient treatment.44 On the other hand, even though Multiplex NAATs for the entire group of RVs, which can be used as POCTs, possess high levels of sensitivity and specificity, they are also very expensive, which limits their routine use, and they are normally reserved for highly specific situations such as critically ill patients admitted to ICUs.43
All in all, given the setting in which we find ourselves, the epidemiological situation and the severity of the clinical symptoms, there is clearly a need to establish diagnostic algorithms to facilitate the rational and efficient use of the different diagnostic tests for RVs (Fig. 2).45,46
Rapid diagnosis of infection by Pneumocystis jirovecii (P. jirovecii)P. jirovecii is a pathogenic fungus that reproduces extracellularly in the pulmonary alveoli of mammals. It does not lead to illness and the immune system remains unaltered. Nonetheless, in situations of immunodepression, Pneumocystis is capable of proliferating and causing severe pneumonia in the absence of specific treatment. As is the case with other colonising pathogens, the detection of P. jirovecii in a respiratory sample is at times difficult to interpret, since its detection does not necessarily imply that it is the agent responsible for the disease.
Pneumocystis spp. has a biphasic life cycle, with two highly distinguished morphological forms: an asexual trophic form, represented by haploid trophozoites, which is the proliferative state and which is fairly abundant; and a cystic form, which represents the reproductive stage, less abundant than the trophic forms, and which is the form of transmission.
Infection by P. jirovecii is difficult to diagnose given that it is a non-culturable fungus. For this reason, the existing diagnostic tests can be classified as rapid as they can be performed in a matter of minutes or a few hours. They can essentially be grouped into staining or molecular techniques, fundamentally PCR, either conventional or real-time.
One important aspect in the diagnosis of Pneumocystis infection is the sample used, which must be combined with the sensitivity of the test employed. Given that Pneumocystis lives in alveolar cells, the pathogenic load can be expected to be greater in samples collected in alveolar cavities, as in bronchoalveolar lavage (BAL) or bronchoalveolar aspirate, than in other samples, such as sputum (either induced or expectorated) and bronchial brushing, or even in samples from the upper respiratory tract, such as oral washings, nasopharyngeal aspirate and nasal swabs.47
Microscopy techniques are generally less sensitive than PCR; hence, it is advisable to always perform them on bronchoalveolar lavage or aspirate. Moreover, these samples are centrifuged before staining in order to concentrate the Pneumocystis. Other respiratory samples in which it can be detected include the fluid from tracheal suction, cell material obtained from bronchial brushing and tissue obtained from transtracheal biopsy.
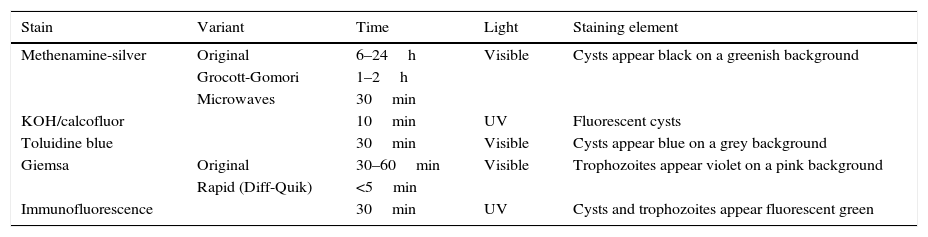
Concerning staining types,48–50 those which have classically been used in diagnosis of infection by Pneumocystis are described in Table 1. By means of methenamine-silver staining, which also stains other fungi, the cysts of Pneumocystis show up black against a greenish background. The greatest drawback is the length of the staining, since the normal procedure may take between 6 and 24h, 1–2h for the abbreviated Grocott-Gomori method or it can be reduced to 30min if performed in a microwave oven. KOH/calcofluor or calcofluor white staining is performed in around 10min, and under fluorescent lights the fungi show up fluorescent blue. Modified toluidine blue staining turns Pneumocystis cysts violet in around 30min.
Most common stains for the diagnosis of pneumonia by Pneumocystis jirovecii in respiratory samples.
| Stain | Variant | Time | Light | Staining element |
|---|---|---|---|---|
| Methenamine-silver | Original | 6–24h | Visible | Cysts appear black on a greenish background |
| Grocott-Gomori | 1–2h | |||
| Microwaves | 30min | |||
| KOH/calcofluor | 10min | UV | Fluorescent cysts | |
| Toluidine blue | 30min | Visible | Cysts appear blue on a grey background | |
| Giemsa | Original | 30–60min | Visible | Trophozoites appear violet on a pink background |
| Rapid (Diff-Quik) | <5min | |||
| Immunofluorescence | 30min | UV | Cysts and trophozoites appear fluorescent green |
In order to observe the trophic forms, the Giemsa stain, which takes around 30–60min, or the rapid variants of Giemsa stain (Diff-Quik), which can be performed in around 5min, are generally used. As samples generally contain around 10 times more trophic forms than cysts, the sensitivity of this staining may be greater than those which only stain cysts.
Pneumocystis can also be detected in around 30min by means of DIF, using monoclonal antibodies targeting surface glycoproteins. Depending on the monoclonal antibody selected, it is possible to specifically stain the cysts or all forms of the microorganism. Given that there are more trophic forms, the sensitivity of those kits which stain both forms is greater. Antibodies are usually marked with fluorescein isothiocyanate, which means that under the ultraviolet light of the microscope, the walls of the cysts and trophozoites show up fluorescent green. While the specificity of all microscopic techniques is similar, the most sensitive is DIF.47,51,52
The most widely used molecular technique is PCR, which is considered to be around 100 times more sensitive than microscopy in the diagnosis of pneumonia caused by P. jirovecii.53 The sensitivity of PCR can be enhanced when using targets present in multiple copies of the Pneumocystis genome, such as the codified gene of the 23S subunit of the mitochondrial rRNA (mitochondrial large subunit rRNA) or the major surface glycoprotein or by performing a nested PCR. Other targets used in the detection of Pneumocystis include the internal transcribed spacer of ribosomal genes, the 5S cytoplasmic rRNA, dihydrofolate reductase and thymidylate synthase.54,55
The high sensitivity of PCRs, capable of detecting quantities lower than 3.5copies/μl of sample, mean that a negative PCR text in a pulmonary sample (BAL, biopsy) serves to rule out the involvement of P. jirovecii in the infection.56
In fact, the high sensitivity of PCR has led some authors to propose it as a diagnostic technique in respiratory samples in which a low quantity of Pneumocystis can be expected, such as samples from immunodepressed patients not infected by HIV or non-invasive respiratory samples (induced sputum, nasal swabs, etc.), since a negative result will avoid the need to resort to invasive techniques. The problem is that in these non-invasive samples, a positive result may detect a sub-clinical colonisation instead of an acute infection. The joint conduct of PCR and microscopy in induced sputum or in samples from the upper respiratory tract has been suggested, such that a positive PCR with a positive microscopy would be a sign of infection, while a positive PCR with negative microscopy would represent colonisation. A negative PCR result would exclude infection and colonisation by Pneumocystis.57
The drawback of the conventional PCR and of the nested PCR is the lack of quantification, the determination of which would be a reflection of the fungal load present and could thus help to differentiate between colonisation and acute infection. This problem can be overcome with real-time quantitative PCR; nonetheless, clear cut-off points for distinguishing between colonisation and illness have yet to be established. In general, a threshold cycle, or an elevated initiation cycle for the exponential phase of the amplification, represents a low fungal load and is possibly more suggestive of a colonisation than an infection.57 In these situations of a positive PCR, serological tests can be added (such as the detection in serum of 1–3 beta-d-glucan), which, even without being specific to Pneumocystis, increases in fungal infections and can supplement a positive PCR result in a non-invasive respiratory sample (sputum or even saliva).58 The negative predictive value of the beta-d-glucan assay is extremely high, and it can be employed in patients on whom bronchoscopy cannot be performed or for whom there is a slight suspicion of infection by Pneumocystis. Reduction in the DNA load of Pneumocystis by means of real-time PCR has also been used as a treatment response control.
It is not uncommon for a patient to be infected with more than one specific genotype of P. jirovecii, which may hinder the detection of a specific genotype or quantification in a quantitative PCR.
Molecular techniques also offer other “alternatives” to the study of infection by Pneumocystis, including epidemiological and person-to-person transmission studies through “multilocus genotyping”, and even an approximation to resistance to trimethoprim-sulfamethoxazole by analysing mutations in the codifying genes of dihydropteroate synthase and dihydrofolate reductase.59
The serological tests for detecting antibodies to P. jirovecii are useful for performing epidemiological studies, but not in the diagnosis of acute infection.
ConclusionGenerally speaking, RDTs are useful for the diagnosis of many RIs, facilitating a better approach to infection in all its aspects. Currently, RDTs for detecting antigens can be used as POCTs, which is not the case for the majority of nucleic acid amplification tests, which, in turn, show higher percentages of specificity, but above all sensitivity. Improvements in terms of the simplicity of sample processing and in the interpretation of results would seem to make it likely that in the near future the use of nucleic acid amplification tests as POCT for the diagnosis of RIs will become more widespread.
Conflicts of interestThe authors declare that there are no conflicts of interest.
Please cite this article as: Marimón JM, Navarro-Marí JM. Métodos de diagnóstico rápido de las infecciones respiratorias. Enferm Infecc Microbiol Clin. 2017;35:108–115.