Since the first plasmid-encoded colistin resistance gene (mcr-1) was described in Escherichia coli in China,1 9 others mobile colistin resistance genes have been identified (mcr-2 to mcr-10).2 A plasmid carrying mcr-8 was first reported in 2018 but similar mechanisms of action for mcr-8 and mcr-2 indicated that mcr-8 resistance genes may have appeared very early.2 Notably, the mcr-8 gene has been detected in patients from intensive care units as well as livestock2 and mcr-8-bearing plasmids have been found primarily in Klebsiella pneumoniae and Raoultella ornithinolytica. In the current study, an IncFIA plasmid carrying mcr-8 was identified in an ST11K. pneumoniae strain isolated from wastewater in a duckery in Fujian, China. Further genetic analysis of this plasmid and others of the same type from global databases were then compared.
The mcr-8-harboring K. pneumoniae isolate ZZW20 was identified by PCR amplification and sequencing of the 16S rRNA. Susceptibility testing was performed following the CLSI3 and the EUCAST4 methodologies, and this strain was resistant to colistin (MIC 32mg/L) and 6 other antimicrobials (Table S1). The whole genome sequence of ZZW20 was determined using a combination of Illumina (San Diego, CA, USA) and MinION (Nanopore, Oxford, UK) platforms and assembled using Unicycle.5 MLST analyses indicated that this mcr-8-positive K. pneumoniae belonged to the most prevalent clonal type (ST11) that carried virulence factors, that was present in Asia, especially China.6
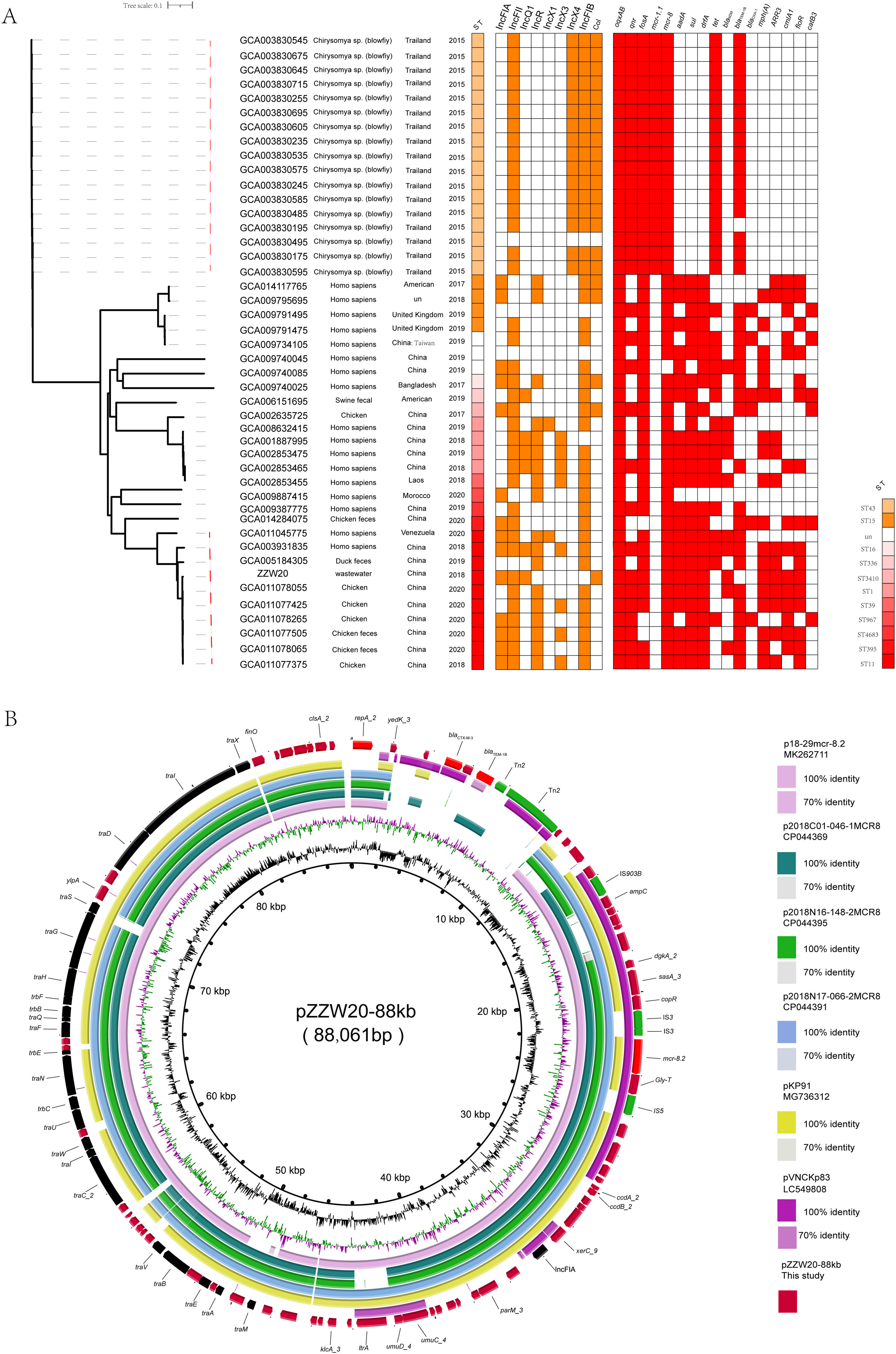
A phylogenetic tree of ZZW20 was generated using 44 additional K. pneumoniae strains carrying mcr-8 on plasmids. The 44 mcr-8-harboring K. pneumoniae strains were primarily from Asia but were distributed within different clades and isolated from humans (n=17) and animals (n=27), indicating a prevalence in animals. A total of 17 blow flies carrying mcr-8 were trapped at three different locations in Northern Thailand: a local market in an urban community, a rural area and a suburb of the city Phitsanulok. Importantly, they were grouped in a cluster, suggesting that flies are likely to be one of the key vectors of the mcr-8. Compared with other strains carrying mcr-8, their ST type was different, indicating that the strains carrying mcr-8 began to evolve toward diversification. Almost all strains of ST11 from China shared 6588 core-genome single nucleotide polymorphisms (cgSNP). Interestingly, significantly distinct cgSNPs (range 1868–4216) were observed between ZZW20 and other database strains suggesting that they did not share a common transmission event (Fig. 1A).
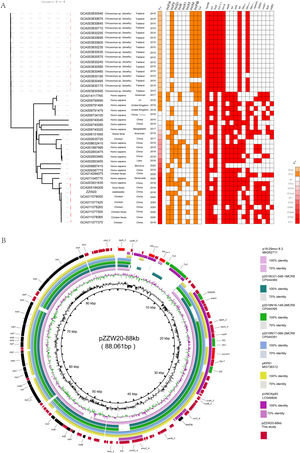
(A) Midpoint-rooted tree generated from the core-genome sequences of the mcr-8-positive ST11K. pneumoniae identified in this study as well as another 43 mcr-8-positive K. pneumoniae isolates. The collection dates, hosts and country of isolation are indicated for each isolate. Major antibiotic resistance genes, ST carried by the K. pneumoniae isolate are indicated. (B) Comparative ananlysis of mcr-8-positive plasmid identified in this study. The circular plasmid map was generated using BRIG. Arrows indicate orientation of open reading frames. Regions of homology are marked by shading.
The mcr-8 gene was present in strain ZZW20 on an ∼88kb plasmid (pZZW20_80K; Acc. No. CP058962) that was identified by S1-PFGE and southern blotting7 using the appropriate probes (Figure S1), and was typed as IncFIA using the WGS data. BLAST analysis demonstrated that pZZW20_80K had a 129kb backbone closely related to the K. pneumoniae plasmids p18-29mcr-8.2 (MK262711), p2018C01-046-1MCR8 (CP044369), p2018N16-148-2MCR8 (CP044395), p2018N17-066-2MCR8 (CP044391), pVNKp83 (LC549808) and PK91 (MG736312). Interestingly, there were only 44 nucleotides that differed between the plasmids in this group (>99.96% identity). Additionally, even though the 7 IncFIA ST11 mcr-8-positive K. pneumoniae isolates were unique (range 1868–4216 cgSNPs), the mcr-8 plasmids they carried were highly similar, especially for pZZW20_80K with 88kb identity (81% query coverage and 99.9% identity) to mcr-8 plasmid p18-29 (MK262711) from a human isolate in China. Then we used these plasmids to make a circular plasmid map with BRIG for further analysis.8 These plasmids shared the genetic context IS903B-ampC-hp-hp-hp-Giy-T-dgkA-baeS-copR-IS3-mcr-8-Gly-T-IS5. However, only the ZZW20 carried blaCTX-M-3 and blaTEM-1B genes. There were also 2 copies of ΔTn2 downstream of blaTEM-1B as well as an intact Tn2 in pZZW20_88K indicating that the insertion of blaCTX-M-3 and blaTEM-1B were most likely mediated by Tn29 (Fig. 1B).
The pZZW20_80K plasmid also possessed the tra/trb gene cluster that mediates plasmid conjugative functions and we therefore conducted conjugation experiments to examine its potential transferability using E. coli EC600str as recipient. The transconjugants that were isolated retained most of the antibiotic resistance genes from the plasmid including colistin resistance (Table S1). This was an additional confirmation that IncFIA plasmids carrying mcr-8 may spread among K. pneumoniae strains.
The 6 virulence factors present on the chromosome of strain ZZW20 were identified using the VFDB database (http://www.mgc.ac.cn/VFs/main.htm). The genes necessary for enterobactin synthesis, transport and utilization (entABCDEF, fepABCDG, fes) as well as the gene responsible for salmochelin cleavage (iroE) were present on the plasmid. The enterobactin siderophore system is the primary iron-sequestering system for Enterobacterales. The presence of salmochelin is associated with invasive disease isolates and is common in highly virulent K. pneumoniae isolates.6 Strain ZZW20 also carried a Type VI Secretion System (T6SS) cluster that contributes to bacterial competition, cell invasion and in vivo colonization that can sensitize hosts to potentially fatal infections by other bacterial pathogens.10 These characteristics of ZZW20 indicated a likelihood that it was evolving into a hypervirulent strain.
In conclusion, ST11K. pneumoniae strains possessing mcr-8 IncFIA plasmids have been sporadically disseminated in China. These plasmids were highly similar and pZZW20_80K -mcr-8 and were able to transfer to E. coli by conjugation. The presence of the highly efficient enterobactin system along with 5 other known virulence factors indicated a potential threat to public health. In particular, we found multiple copies of Δ IS66 transposases on the mcr-8 plasmid revealing the potential transferability of mcr-8. Thus, there is an urgent need for further surveillance to understand the prevalence and dissemination of mcr-8-positive K. pneumoniae and prevent future disease outbreaks.
Accession number. Sequence of pZZW20-88K has been assigned GenBank accession number CP058962.
FundingThis work was supported by grants from the National Key Research and Development Program of China (2016YFD0501300), the Program of Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (IRT_17R39), the Guangdong Special Support Program Innovation Team (2019BT02N054), the Foundation for Innovation and Strengthening School Project of Guangdong, China (2016KCXTD010) and the Innovation Team Project of Guangdong University (2019KCXTD001).