This study reports the emergence of mcr-1-mediated colistin resistance in human Escherichia coli isolates from Bulgaria.
MethodsThree colistin-resistant E. coli isolates were obtained from outpatient urine specimens. They were subjected to PCR for detection of mcr genes and conjugation experiments. Whole-genome sequencing was employed to analyze the genomic characteristics of the isolates.
ResultsPCR identified mcr-1 in all isolates. In E. coli of sequence type (ST) 2067, mcr-1.1 was found on a self-transmissible IncI2 plasmid, while mcr-1.32 was chromosomal in the remaining two ST131 E. coli isolates. E. coli ST2067 co-harbored quinolones resistance mutations (gyrAD87N, gyrAS83L, parCS80I), β-lactam (blaTEM-30) and aminoglycoside (aadA1, aac(3)-IId) resistance genes.
ConclusionThis report further confirms the role of IncI2 conjugative plasmids in the dissemination of mcr genes. Our findings involving chromosomal mcr-1 in high-risk ST131 E. coli strains from outpatients underscores the need for enhanced surveillance and systematic screening to combat colistin resistance.
En este estudio se informa de la aparición de resistencia a la colistina mediada por mcr-1 en cepas humanas de Escherichia coli procedentes de Bulgaria.
MétodosSe obtuvieron tres aislados de E. coli resistentes a la colistina a partir de muestras de orina de pacientes ambulatorios. Se sometieron a PCR para la detección de genes mcr y a experimentos de conjugación. Se empleó la secuenciación del genoma completo para analizar las características genómicas de los aislados.
ResultadosLa PCR identificó mcr-1 en todos los aislados. En E. coli de tipo de secuencia (ST) 2067, mcr-1.1 se encontró en un plásmido IncI2 autotransmisible, mientras que mcr-1.32 era cromosómico en los dos aislados restantes de E. coli ST131. E. coli ST2067 albergaba mutaciones de resistencia a quinolonas (gyrAD87N, gyrAS83L, parCS80I), a β-lactámicos (blaTEM-30) y a aminoglucósidos (aadA1, aac(3)-IId).
ConclusionesEste informe confirma aún más el papel de los plásmidos conjugativos IncI2 en la diseminación de genes mcr. El hallazgo de mcr-1 cromosómico en cepas de E. coli ST131 de alto riesgo procedentes de pacientes ambulatorios subraya la necesidad de mejorar la vigilancia y el cribado sistemático para combatir la resistencia a la colistina.
Colistin has been recognized as a last resort antibiotic for treating infections caused by multi-resistant Gram-negative bacteria. However, in 2016, Liu et al. described the first plasmid-mediated colistin resistance gene (mcr-1) found in Escherichia coli from animal and human samples, and later its emergence was tracked back to the 1980s. MCR enzymes modify lipid A through phosphoethanolamine transfer blocking the attachment of colistin and conferring resistance or reduced susceptibility.1
Because of the increasing resistance to third-generation cephalosporins, fluoroquinolones and aminoglycosides, carbapenems have become the preferred antibiotics for the treatment of complicated infections. However, this has led to the emergence and spread of carbapenem-resistant E. coli, which often have a multidrug-resistant phenotype, requiring treatment with β-lactam–β-lactamase inhibitor combinations (e.g. ceftazidime–avibactam, meropenem–vaborbactam, imipenem–cilastatin–relebactam), cefiderocol, or combination therapy with other drugs, including colistin.2,3 The primary obstacle to colistin-based therapy, in addition to its toxic nature, is the rapid surge in colistin resistance. The plethora of chromosomal and often intricate resistance mechanisms have been succeeded by mobile colistin resistance (mcr) genes, frequently detected among colistin-resistant E. coli.1 Existing studies have shown that mcr genes are usually located on plasmids of IncI2, IncX4, IncP, IncX and IncFII types, and their spread is aided by insertion sequences (IS) such as ISApl1 and transposon Tn6330.1,4 Although rare, chromosomally-encoded mcr genes have also been reported.5
The aim of this study was to investigate the genomic characteristics of three clinical E. coli strains harboring the mcr-1 gene, which was plasmid-encoded in one isolate and chromosomal in two of them. As far as we are aware, this is the first report of mcr-1-positive bacteria from human sources in Bulgaria.
Material and methodsBacterial isolation, identification, antimicrobial susceptibility testing and conjugation experimentsThe three isolates (EC707, EC2947, EC1752) were recovered from urine specimens of outpatients presented with community-acquired dysuria in the primary care unit of a cancer hospital. Isolate EC707 was collected in March 2022 from a 38-year-old woman, EC2947 – in October 2022 from a 56-year-old woman, and EC1752 – in May 2023 from a 46-year-old woman.
Isolates were identified by MALDI-TOF Biotyper (Bruker Daltonics GmbH, Bremen, Germany) with MALDI Reference 2022 Library v.4.0.
Colistin resistance was initially screened with SuperPolymyxin medium,6 which was performed on all 2342 E. coli isolates recovered from patients’ specimens between January 2017 and December 2023. Clinical specimens were obtained from infected or colonized patients admitted to hospital wards or attending outpatient departments of a 252-bed oncology hospital in Sofia, Bulgaria.
Colistin resistance of the three screened isolates, which represented 0.1% of all isolates tested, was then confirmed by broth microdilution with MIC-strip colistin (Bruker Daltonics GmbH, Bremen, Germany). The minimum inhibitory concentrations (MICs) of a wide range of antimicrobials were also determined using the MicroScan NM-EN52 panel (Beckman Coulter, Inc., Brea, CA, USA) following the manufacturer's protocol. Susceptibility to fosfomycin was determined by the disk diffusion method on Mueller–Hinton agar with disks supplied by Becton Dickinson (BD, Sparks, MD, USA). The results of susceptibility testing were interpreted according to EUCAST clinical breakpoints v14.0. E. coli ATCC 25922 and E. coli NCTC 13846 were used for quality control.
The transmissibility of colistin resistance was tested by mating with sodium azide-resistant E. coli J53 on Luria-Bertani agar plates containing 150mg/L sodium azide and 2mg/L colistin sulphate. Transconjugants were confirmed by susceptibility testing and PCR.
Screening for mcr genesTotal genomic DNA for PCR and whole-genome sequencing (WGS) was extracted using the PureLink™ Genomic DNA Mini Kit (Thermo Fisher Scientific, Missouri, TX, USA) with all homogenization steps carried out by pipetting.
Multiplex PCR for detection of mcr genes was performed as described previously.7
Whole-genome sequencing and bioinformatic analysisLong-read sequencing was performed on MinION Mk1C with the Rapid Barcoding Kit 96 (SQK-RBK110.96) and FLO-MIN106D (R9.4.1) (Oxford Nanopore Technologies, Oxford, UK).
Read filtering was performed with Filtlong v0.2.1 (https://github.com/rrwick/Filtlong, accessed 6.12.23). Assemblies were produced with Trycycler v0.5.3,8 followed by polishing with MEDAKA v1.7.3 (ONT, https://github.com/nanoporetech/medaka, accessed 21.2.24). Screening for antimicrobial resistance genes was done with AMRFinderPlus v3.11.4.9 Plasmids were analyzed with Abricate (Seemann T, Abricate, Github https://github.com/tseemann/abricate) using the PlasmidFinder database (v2023-01-18).10 Multi-locus sequence typing (MLST) was performed using the Achtman scheme with mlst v2.23.0 (Seemann T, mlst Github https://github.com/tseemann/mlst, accessed 21.2.24). Closely related sequences were retrieved by BLAST using the nr/nt database (Update date: 19 March 2024). Gview11 and Clinker12 were used for visualization.
ResultsPCR revealed a 213bp band characteristic for mcr-1 in all E. coli isolates. Complete genomes were obtained for all three isolates (European Nucleotide Archive accession number PRJEB70793). EC707 had a genome size of 4.9 Mb, carrying three plasmids of sizes 154kb (with IncFIB and IncFIC replicons), 110kb (with IncI1-I(Alpha) replicon), and 64kb (IncI2 replicon). Similarly, EC1752 and EC2947 had genome sizes of ∼4.97Mb and ∼4.99Mb, respectively. Each of them possessed two nearly identical plasmids of sizes 125kb (with IncFIB and IncFII replicons), and 6.6kb (untypable).
In silico MLST identified EC707 as sequence type (ST) 2067, while both EC1752 and EC2947 were ST131.
WGS analysis of EC707 revealed mutations in genes associated with resistance to quinolones (gyrAD87N, gyrAS83L, parCS80I), fosfomycin (glpTE448K), and colistin (pmrBY358N). Detected resistance genes included mcr-1.1, tet(A), aadA1, aac(3)-IId and the inhibitor-resistant broad-spectrum blaTEM-30.
EC1752 and EC2947 had a similar resistance profile, characterized by mutations in genes associated with fosfomycin (glpTE448K, uhpTE350Q), colistin (pmrBE123D), and quinolone (parEI529L) resistance, along with the mcr-1.32 gene. Interestingly, all resistance determinants in these two strains were chromosomally encoded.
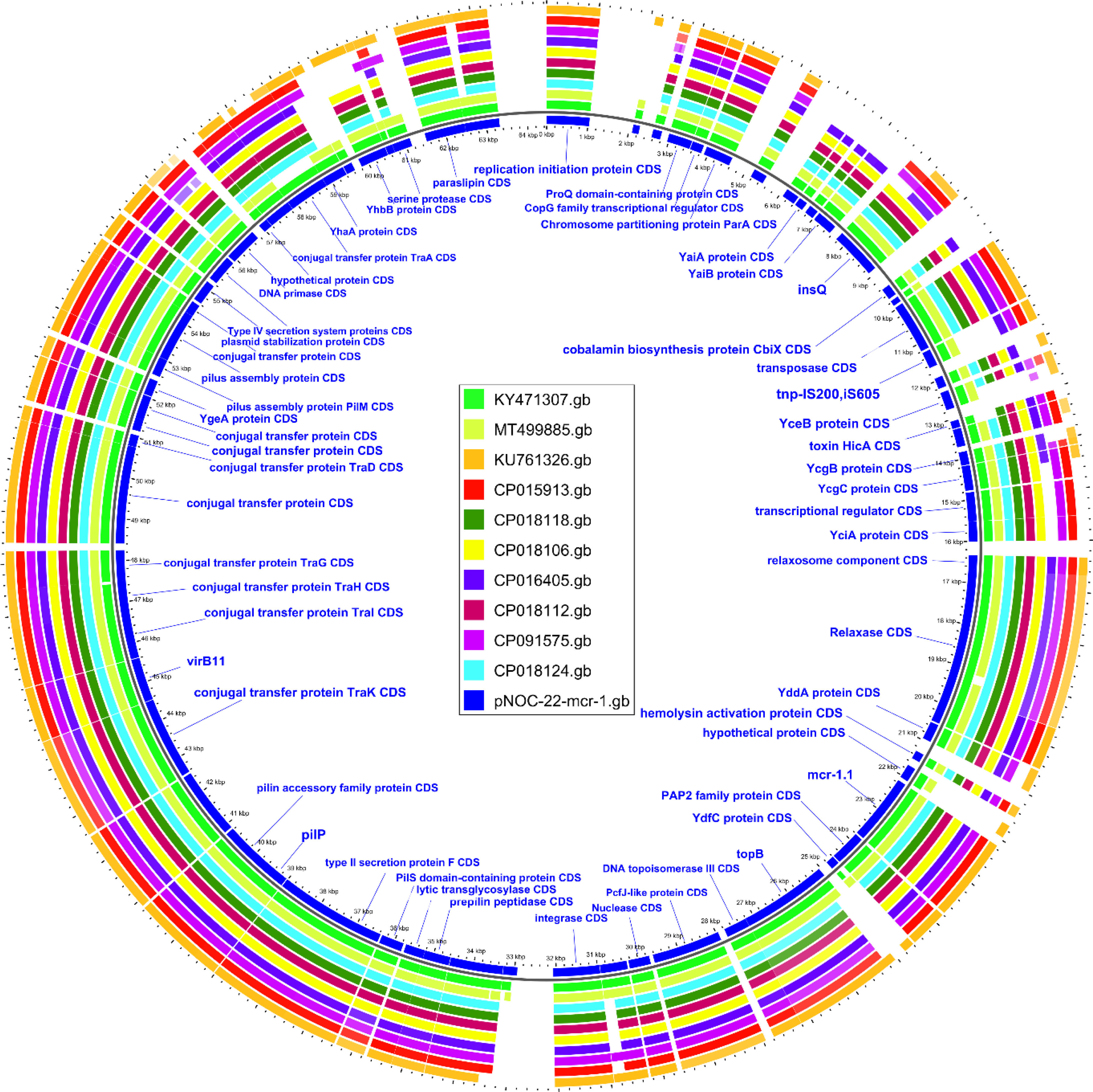
The mcr-1.1 variant in EC707 was located on the 64kb IncI2 plasmid (pNOC-22-mcr-1). The ten genetically closest plasmids were retrieved from NCBI through BLAST analysis, and compared with pNOC-22-mcr-1 (Fig. 1). A 99.9% identity was found between pNOC-22-mcr-1 and an IncI2 plasmid (pMCR-GN775; KY471307) from a Canadian isolate belonging to ST624.13
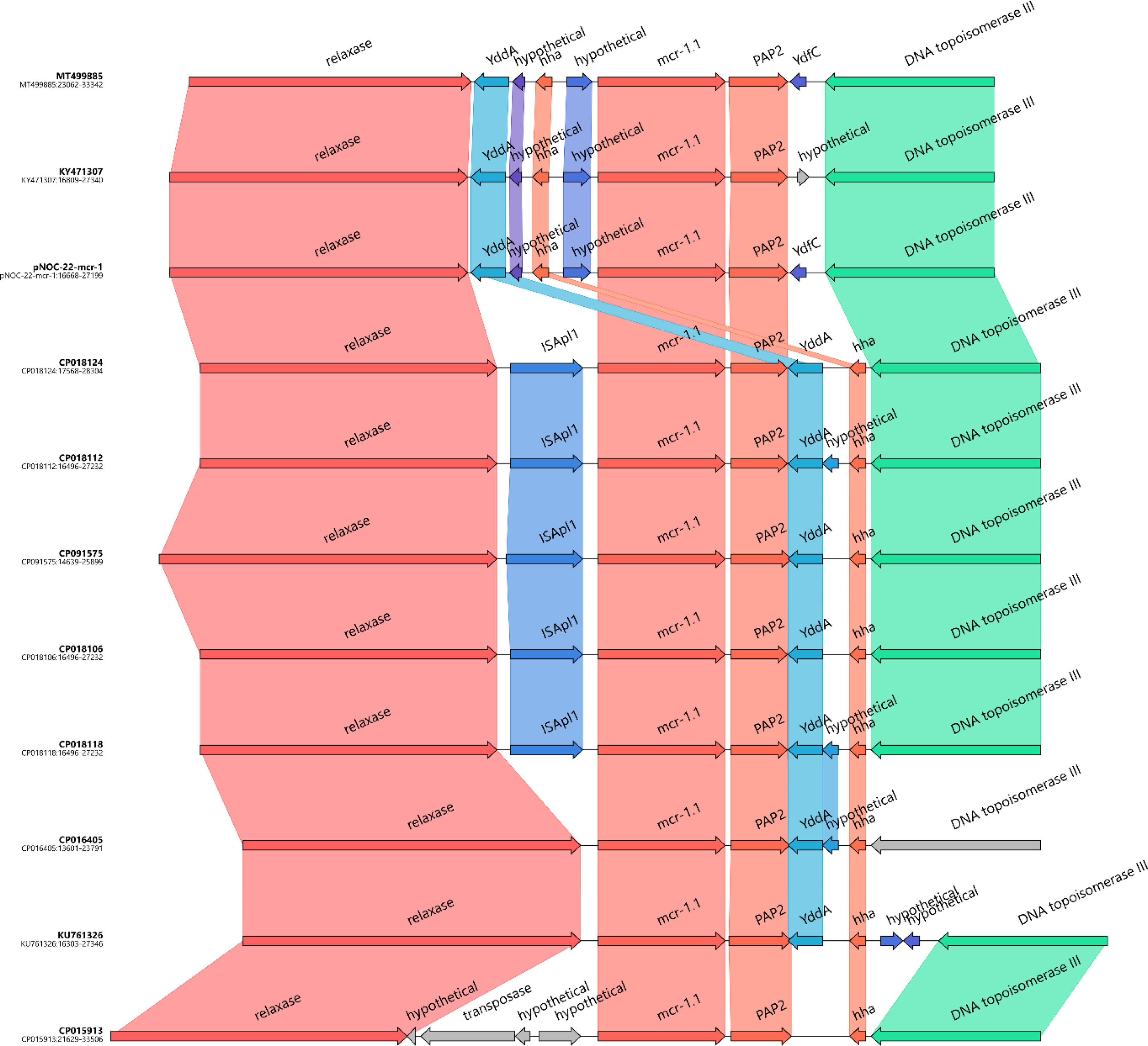
Analysis of the genetic structures, which surrounded the mcr-1.1 segment containing the PAP2 gene, revealed the lack of ISApl1 similar to that of pMCR-GN775 (KY471307) (Fig. 2), and of two IncI2 plasmids previously described in isolates from Colombian hospitals, and belonging to ST58 and ST46.14
In EC1752 and EC2947, the segment containing mcr-1.32 and PAP2 genes was chromosomally located within a truncated IS66. Remnants of this IS were found both upstream and downstream of the mcr segment. Besides the IS66 remnant downstream of PAP2, the region consisted of IS110 followed by a gene encoding lactate dehydrogenase. Upstream of mcr-1.32, the region included the remaining IS66 remnant, followed by two genes – one encoding a hypothetical protein and the other encoding a GTPase family protein. The same genetic structure was previously described in mcr-1.13-positive E. coli isolates from turkeys and pigs in Italy.15
Consistent with the WGS results, a colistin-resistant transconjugant was obtained from EC707 by conjugal transfer of the IncI2 plasmid carrying only mcr-1.
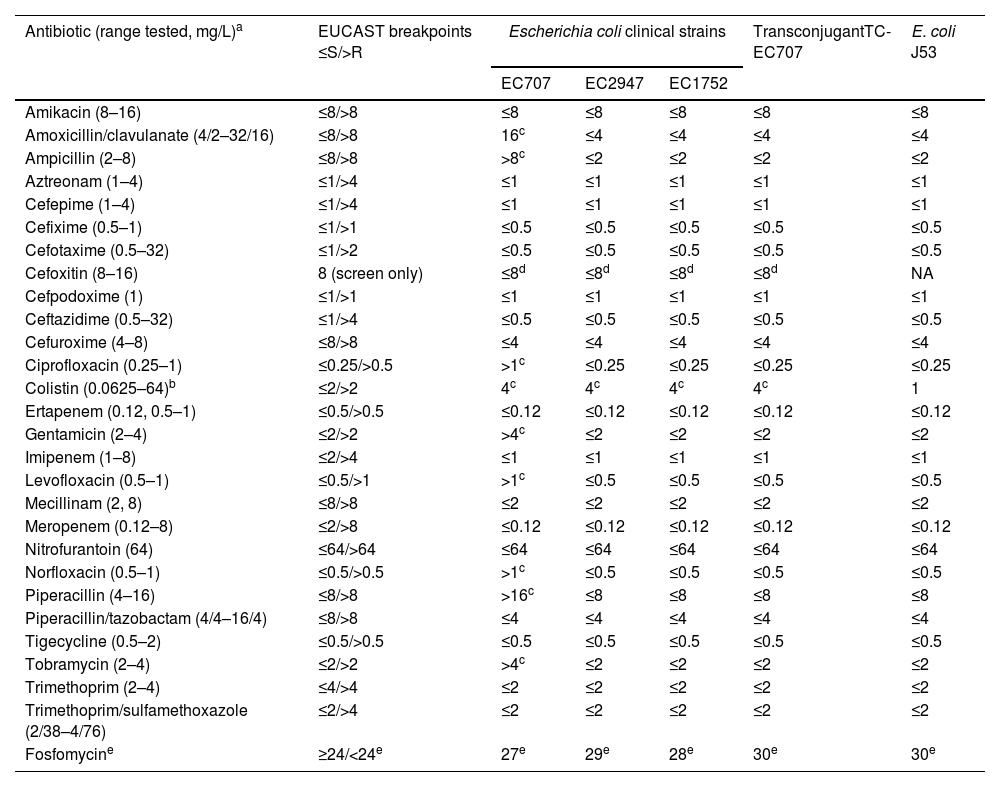
All the three isolates, as well as the transconjugant TC-EC707, exhibited resistance to colistin with MIC of 4mg/L due to mcr genes (Table 1). EC1752 and EC2947 remained susceptible to all other antimicrobials tested, while EC707 showed resistance to penicillins and their inhibitor combinations, to gentamicin and tobramycin, and to quinolones due to blaTEM-30, to aac(3)-IId, and to mutations in gyrA and parC, respectively. Long-read sequencing of EC707 revealed that blaTEM-30 was located on the IncI1-I(Alpha) plasmid, which also carried the aac(3)-IId acetyltransferase gene.
Antimicrobial susceptibility of mcr-1-positive Escherichia coli clinical strains, transconjugant TC-EC707 and E. coli J53 recipient strain.
| Antibiotic (range tested, mg/L)a | EUCAST breakpoints ≤S/>R | Escherichia coli clinical strains | TransconjugantTC-EC707 | E. coli J53 | ||
|---|---|---|---|---|---|---|
| EC707 | EC2947 | EC1752 | ||||
| Amikacin (8–16) | ≤8/>8 | ≤8 | ≤8 | ≤8 | ≤8 | ≤8 |
| Amoxicillin/clavulanate (4/2–32/16) | ≤8/>8 | 16c | ≤4 | ≤4 | ≤4 | ≤4 |
| Ampicillin (2–8) | ≤8/>8 | >8c | ≤2 | ≤2 | ≤2 | ≤2 |
| Aztreonam (1–4) | ≤1/>4 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| Cefepime (1–4) | ≤1/>4 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| Cefixime (0.5–1) | ≤1/>1 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Cefotaxime (0.5–32) | ≤1/>2 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Cefoxitin (8–16) | 8 (screen only) | ≤8d | ≤8d | ≤8d | ≤8d | NA |
| Cefpodoxime (1) | ≤1/>1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| Ceftazidime (0.5–32) | ≤1/>4 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Cefuroxime (4–8) | ≤8/>8 | ≤4 | ≤4 | ≤4 | ≤4 | ≤4 |
| Ciprofloxacin (0.25–1) | ≤0.25/>0.5 | >1c | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Colistin (0.0625–64)b | ≤2/>2 | 4c | 4c | 4c | 4c | 1 |
| Ertapenem (0.12, 0.5–1) | ≤0.5/>0.5 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 |
| Gentamicin (2–4) | ≤2/>2 | >4c | ≤2 | ≤2 | ≤2 | ≤2 |
| Imipenem (1–8) | ≤2/>4 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| Levofloxacin (0.5–1) | ≤0.5/>1 | >1c | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Mecillinam (2, 8) | ≤8/>8 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 |
| Meropenem (0.12–8) | ≤2/>8 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 |
| Nitrofurantoin (64) | ≤64/>64 | ≤64 | ≤64 | ≤64 | ≤64 | ≤64 |
| Norfloxacin (0.5–1) | ≤0.5/>0.5 | >1c | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Piperacillin (4–16) | ≤8/>8 | >16c | ≤8 | ≤8 | ≤8 | ≤8 |
| Piperacillin/tazobactam (4/4–16/4) | ≤8/>8 | ≤4 | ≤4 | ≤4 | ≤4 | ≤4 |
| Tigecycline (0.5–2) | ≤0.5/>0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Tobramycin (2–4) | ≤2/>2 | >4c | ≤2 | ≤2 | ≤2 | ≤2 |
| Trimethoprim (2–4) | ≤4/>4 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 |
| Trimethoprim/sulfamethoxazole (2/38–4/76) | ≤2/>4 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 |
| Fosfomycine | ≥24/<24e | 27e | 29e | 28e | 30e | 30e |
MIC values were determined using the MicroScan NM-EN52 panel (Beckman Coulter, Inc., Brea, CA, USA).
The current study describes the first mcr-positive E. coli isolates of human origin in Bulgaria. Unlike most publications,4 colistin-resistant isolates in this study were collected only from outpatients. Notably, we observed chromosomal localization of the mcr-1.32 gene in two isolates, which appears less common compared to the plasmid one. These isolates belonged to the high-risk E. coli clone ST131, which was frequently associated with CTX-M-15 production and fluoroquinolone resistance. Fluoroquinolone/cephalosporin-resistant E. coli isolates from this clone often carry additional resistance determinants and virulence genes, and are associated with a wide range of infections.16 Furthermore, we identified mcr-1.1 in EC707 (ST2067) on a self-transferable IncI2 plasmid, which has been involved in the dissemination of mcr-1 gene.13
Previous studies have associated pmrBY358N and pmrBE123D mutations with colistin resistance. However, their impact remains uncertain.17 In our study, the presence of mcr genes obscured their contribution to the observed colistin MIC.
The detection of mcr genes among outpatients raises concerns, considering that colistin susceptibility is not routinely tested in outpatient settings. Therefore, the true extent of mcr prevalence in Bulgaria could be underestimated, especially since the zoonotic prevalence is still unidentified. We were able to identify the mcr-positive isolates through regular colistin resistance screening performed on all isolates in our laboratory. This emphasizes the importance of systematic surveillance and the incorporation of a targeted screening for colistin resistance for all isolates to mitigate the spread of resistance.
FundingThis research was funded by the European Regional Development Fund through the Operational Program Science and Education for Smart Growth 2014–2020; Grant BG05M2OP001-1.002-0001-C04 “Fundamental Translational and Clinical Research in Infection and Immunity”.
Conflicts of interestNone.