The objective of this study was to assess the performance of a technique (S. PneumoStrip test) based on PCR followed by reverse strip hybridisation for the detection of Streptococcus pneumoniae serotypes directly in blood culture vials.
MethodsOne hundred and ten (110) pairs of isolated strains and their corresponding original blood cultures vials were studied in parallel. Pure isolated strains were conventionally serotyped using latex agglutination and the Quellung reaction. The S. PneumoStrip test was carried out directly in the original blood culture samples.
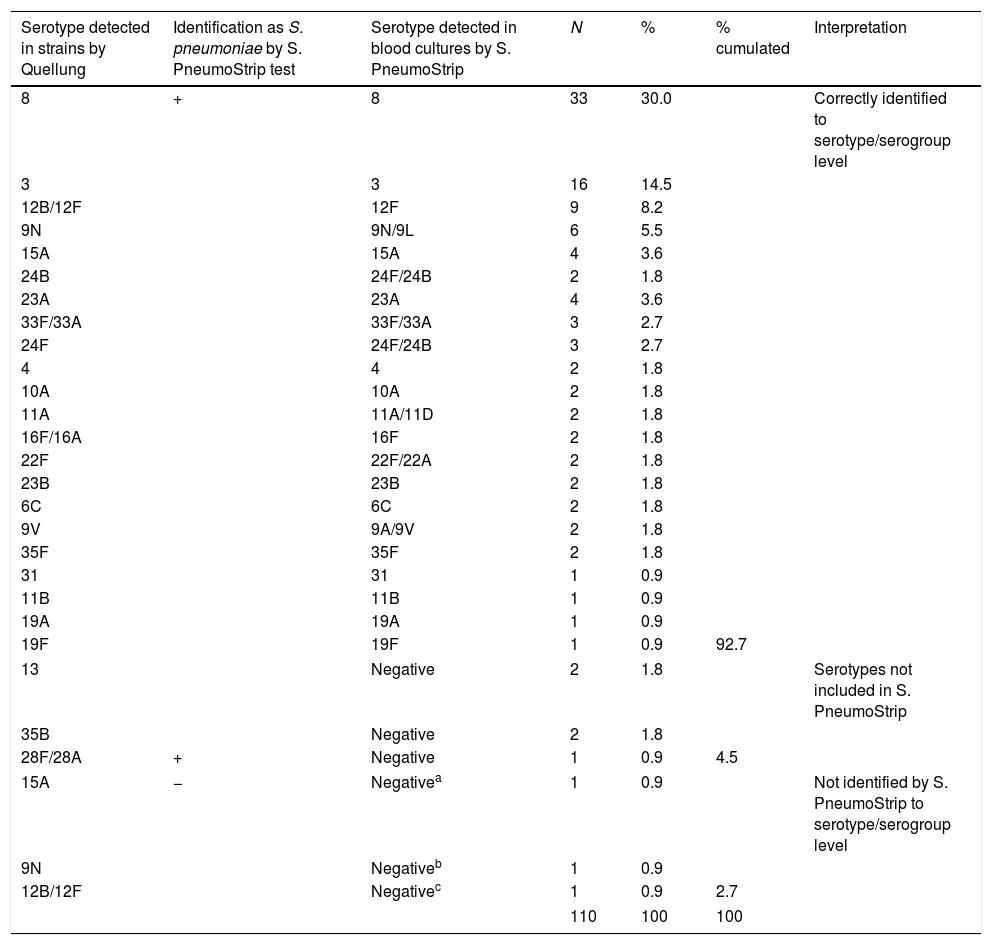
ResultsIn 102 cases (92.7%), results of the serotype obtained by Quellung coincided with their corresponding original blood cultures typed by S. PneumoStrip.
ConclusionsS. PneumoStrip test is a good alternative technique for direct pneumococcal serotyping in blood culture clinical samples.
El objetivo de este estudio fue evaluar el rendimiento de una técnica de PCR seguida de hibridación reversa (S. PneumoStrip test) para su aplicación directa en muestras de frasco de hemocultivo.
MétodosSe estudiaron 110 cepas aisladas en sangre y sus correspondientes muestras de frasco de hemocultivo. Las cepas fueron serotipadas convencionalmente mediante aglutinación por látex y reacción de Quellung. El test de S. PneumoStrip se realizó directamente en las muestras originales de hemocultivo.
ResultadosEn 102 casos (92,7%) el serotipado por Quellung coincidió con el obtenido directamente en su original frasco de hemocultivo.
ConclusionesS. PneumoStrip constituye una buena alternativa para el serotipado directo en muestras de frasco de hemocultivo.
Streptococcus pneumoniae serotyping remains critical in the epidemiological surveillance of invasive pneumococcal disease (IPD). The Quellung reaction is the gold standard for serotyping pneumococci. In recent years, some alternative serotyping procedures that include PCR, real-time PCR, sequencing analysis of specific genes, and whole genome sequencing, have been developed.1 Some of these techniques are reduced to a limited number of detectable serotypes2 or require expensive equipment (sequencing analysis).1 Recently, a commercial assay “S. PneumoStrip test” (Operon S.A., Zaragoza, Spain) for pneumococcal serotyping in strains isolated in culture, based on PCR followed by reverse strip hybridisation, has been reported as easy, sensitive and specific procedure.3 The targets of the test are 76 serotypes (42 individually and 34 in pairs). The objective of this work was to assess the performance of the “S. PneumoStrip test” for the detection of S. pneumoniae serotypes directly in blood culture vials.
Materials and methodsOne hundred and ten (110) pairs of isolated strains and their corresponding original blood cultures vials, consecutively collected in three public hospitals within the Community of Madrid between July 2017 and July 2018, were studied in parallel (five corresponded to paediatric cases and the rest were from adult patients). Pure isolated strains were conventionally serotyped using the ImmuLex™ Pneumotest Kit (Statens Serum Institut, Copenhagen, Denmark), and the Quellung test using pneumococcal factor anti-sera (Statens Serum Institut, Copenhagen, Denmark) was used to complete the full serotyping of the strains.
The S. PneumoStrip test was carried out directly in blood culture samples. For DNA bacterial extraction, 5mL of blood culture vials were centrifuged at 1000 revolutions per minute (rpm) for 2min and the supernatants were subsequently re-centrifuged at 10,000rpm for 3min. The pellet was used for DNA isolation and purification by using an automated magnetic particle technology with the Qiagen kit (kit EZ1® DSP Virus; Hilden, Germany).
Amplifications were performed according to the S. PneumoStrip manufacturer's recommendations using an ABI 7500 Fast instrument (Applied Biosystems, Foster City, California, USA). The denatured amplicons were placed in three hybridisation nylon strips that include 76 target serotypes among the three strips. Strip A contains the autolysin (lytA) and the capsular (cpsA) genes and serotypes 1, 3, 4, 5, 6A, 6B, 6C, 6D, 7F/7A, 9A/9V, 14, 18A, 18B/18C, 18F, 19A, 19F, 23A, 23B and 23F. Strip B includes the pneumolysin (ply) gene and serotypes 2, 8, 9N/9L, 10A, 10B, 10F/10C, 11A/11D, 11B, 11C, 11F, 12A/46, 12B/44, 12F, 15B/15C, 17F, 20, 22F/22A, 33F/33A and 37. Finally, strip C again includes the lytA gene and serotypes 7B, 7C/40, 15A, 15F, 16F, 19B/19C, 21, 25A/25F, 38, 24A, 24B/24F, 31, 32A/32F, 33B/33D, 33C, 35A, 35C, 35F/47F, 41A and 41.3
ResultsThe serotypes identified by Quellung from the 110 studied strains belonged to 21 single serotypes (8 [30.0%], 3 [14.5%], 9N [6.4%], 15A [4.5%], 23A [3.6%], 24F [2.7%], 4 [1.8%], 10A [1.8%], 11A [1.8%], 13 [1.8%], 22F [1.8%], 23B [1.8%], 24B [1.8%], 35B [1.8%], 35F [1.8%], 6C [1.8%], 9V [1.8%]), 11B [0.9%], 19A [0.9%], 19F [0.9%], and,31 [0.9%], and 4 pairs of serotypes (12/B12F [9.1%], 33F/33A [2.7%], 16F/16A [1.8%], 28F/28A [0.9%] [in these pairs the sera factor cross-reactions cannot allow us to definitively assign a specific serotype microscopically]). Paired strains and blood culture-detected serotypes are shown in Table 1. In 102 cases (92.7%) the serotype results obtained in the direct strain by Quellung and in the corresponding original blood culture with S. PneumoStrip coincided. In 5 cases (4.5%), the serotype detected in the strains by Quellung was not included within the 76 serotypes of the S. PneumoStrip test and, hence, was not identified directly in blood culture. In 3 cases (2.7%), the S. PneumoStrip results in blood culture were negative (no amplification of lytA, cpsA and ply control genes) despite the Quellung reaction identifying serotypes 15A, 9N and 12B/12F in the strains. These 3 strains isolated in agar plates were re-tested by S. PneumoStrip and the obtained results coincided with the Quellung serotype determinations 15A, 9N and 12F. S. PneumoStrip allowed serotype-level identification for some serotypes that cannot be completely differentiated by the Quellung reaction (serotypes 12/B12F classified by S. PneumoStrip as serotype 12F with simultaneously positive bands for 12B/12F/44 and 12F; and serotypes 16F/16A assigned by S. PneumoStrip as 16F by a specific band). Inversely, the serotype 24F identified by Quellung was not differentiated by S. PneumoStrip (24F/24B). The global sensitivity for capsular pneumococci (detection of lytA, cpsA and ply control genes) directly in blood cultures was 107/110 (97.3%). Specificity was 100% confirming an accurate level of serotype discrimination using S. PneumoStrip, with no misidentification directly in blood cultures.
Paired serotypes detected in strains by Quellung and directly in blood culture by S. PneumoStrip Test.
| Serotype detected in strains by Quellung | Identification as S. pneumoniae by S. PneumoStrip test | Serotype detected in blood cultures by S. PneumoStrip | N | % | % cumulated | Interpretation |
|---|---|---|---|---|---|---|
| 8 | + | 8 | 33 | 30.0 | Correctly identified to serotype/serogroup level | |
| 3 | 3 | 16 | 14.5 | |||
| 12B/12F | 12F | 9 | 8.2 | |||
| 9N | 9N/9L | 6 | 5.5 | |||
| 15A | 15A | 4 | 3.6 | |||
| 24B | 24F/24B | 2 | 1.8 | |||
| 23A | 23A | 4 | 3.6 | |||
| 33F/33A | 33F/33A | 3 | 2.7 | |||
| 24F | 24F/24B | 3 | 2.7 | |||
| 4 | 4 | 2 | 1.8 | |||
| 10A | 10A | 2 | 1.8 | |||
| 11A | 11A/11D | 2 | 1.8 | |||
| 16F/16A | 16F | 2 | 1.8 | |||
| 22F | 22F/22A | 2 | 1.8 | |||
| 23B | 23B | 2 | 1.8 | |||
| 6C | 6C | 2 | 1.8 | |||
| 9V | 9A/9V | 2 | 1.8 | |||
| 35F | 35F | 2 | 1.8 | |||
| 31 | 31 | 1 | 0.9 | |||
| 11B | 11B | 1 | 0.9 | |||
| 19A | 19A | 1 | 0.9 | |||
| 19F | 19F | 1 | 0.9 | 92.7 | ||
| 13 | Negative | 2 | 1.8 | Serotypes not included in S. PneumoStrip | ||
| 35B | Negative | 2 | 1.8 | |||
| 28F/28A | + | Negative | 1 | 0.9 | 4.5 | |
| 15A | − | Negativea | 1 | 0.9 | Not identified by S. PneumoStrip to serotype/serogroup level | |
| 9N | Negativeb | 1 | 0.9 | |||
| 12B/12F | Negativec | 1 | 0.9 | 2.7 | ||
| 110 | 100 | 100 | ||||
To date, up to 99 serotypes of pneumococcus have been described based on the capsular Quellung reaction with commercial rabbit antisera and molecular approaches.4,5 Although the Quellung test still remains the conventional standard procedure for pneumococcal serotyping,1,4 it is a labour intensive technique that is time-consuming and involves large panels of antisera. Moreover, an additional limitation of the Quellung test is the fact that it requires pneumococcal isolate growth, and therefore, its sensitivity is greatly affected by the culture method. PCR-based serotyping techniques are faster and less laborious than phenotypic methods.4 However, they are limited by the number of targets covered. S. PneumoStrip would permit the identification of 76 serotypes according to their capsular polysaccharide biosynthesis (cps) locus sequence. It also would be able to detect unencapsulated strains (positive for lytA and ply but negative for cpsA and any serotype band) as well as multiple-serotype infections.6 It would be important in colonisation studies because the presence of multiple pneumococcal serotypes is difficult to recognise with conventional culture and the Quellung reaction methods. Nevertheless, PCR serotyping techniques may not be able to distinguish between closely related serotypes when gene sequences vary by only a few mutations.4 In this sense, some serotypes are identified together by the S. PneumoStrip Scheme.6 In this study, in some cases where the Quellung showed cross-reactions (pairs 12/B12F and 16F/16A), S. PneumoStrip allowed the precise identification of serotype 12F (simultaneously positive bands for 12B/12F/44 and 12F) and 16F (single positive band for 16F). The 13-valent conjugate vaccine (PCV13) covers serotypes 1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19F, 19A, and 23F. Serotypes 22F and 33F are candidates for the new 15-valent pneumococcal conjugate vaccine (PCV15).7,8 The recently patented 20-valent pneumococcal conjugate vaccine (PCV20) adds the five serotypes 8, 10A, 11A, 12F and 15B to the PCV15.9 All these serotypes are included in the S. PneumoStrip technique, with some (7F, 9V, 22F, 33F, 11A, 15B, 18C) paired to other serotypes of the same serogroup (7A, 9A, 22A, 33A, 11D, 15C, 18B).
The distribution of serotypes observed in this study may reflect the circulation of the pneumococcal population in our region. In Madrid, during the period 2013–2015, the most frequent serotypes identified were 8, 3, 19A, 22F, 9N, 11A, 23B and 10A respectively.10 All these serotypes were detected in this work although some were identified by S. PneumoStrip in pairs (9N/9L, 22F/22A, 11A/11D). The non-conjugate covered serotypes 13, 35B and 28F/28A, that were detected by the Quellung test in isolated strains, but not by S. PneumoStrip, are rare (incidence in Madrid from 2013 to 2015 at <0.01 per 100.000).10 In the current study, we found three blood culture positive hits (2.7%) resulting as negative cases with S. PneumoStrip. This discrepancy was not due to S. PneumoStrip failures (the results in the strains coincided with the Quellung) and might be related to an amplification failure due to low bacterial load in the blood culture media or may be due to a weak hybridisation step limiting the optical reading of reactive bands. However, the global sensitivity reached for capsular pneumococci detection and for serotype identification was high, providing complete specificity. In conclusion, the S. PneumoStrip test is a good alternative approach for direct pneumococcal serotyping in blood culture clinical samples. It would allow to detect the most frequent serotypes circulating in our region.
FinancingThe kits of S. PneumoStrip used in this study were provided Operon S.A., Zaragoza, Spain.
Conflict of interestDisclosure: MS. Ruth de Luis, Ms Susana del Río and Ms Susana Gamen; OPERON S.A. (Zaragoza, Spain).