The aim of this study was to compare the population structure of three different representative groups of E. coli isolates causing urinary tract infections in a large area of Madrid, Spain: two groups of multidrug resistant isolates (MDR), ESBL- and non-ESBL producers, and one of fully-susceptible isolates (35 isolates in each group).
MethodsEpidemiological relatedness was studied by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). The presence of genes encoding ESBL was determined by using PCR and sequencing. Antimicrobial susceptibility testing was performed by broth microdilution.
ResultsPFGE analysis revealed a high degree of genetic diversity in susceptible and non-ESBL-MDR groups. However, the ESBL-MDR E. coli population was less diverse and a large cluster consisting of ST131 and CTX-M-15-producing isolates was detected.
ConclusionsThe present study revealed that ESBL-producing-MDR E. coli population was less diverse than the non-ESBL MDR group and that ST131 was dominant among CTX-M-15-producing isolates that reflects the spread of this successful MDR lineage.
El objetivo del presente estudio fue comparar la estructura poblacional de tres grupos representativos diferentes de aislados de E. coli que producen infecciones del tracto urinario en una gran área de Madrid, España: dos grupos de aislados multirresistentes (multidrug resistant, MDR), productores y no productores de BLEE (β-lactamasas de espectro extendido), y uno de aislados totalmente sensibles (35 aislamientos en cada grupo).
MétodosLa relación epidemiológica se estudió mediante electroforesis en gel de campo pulsado (pulsed-field gel electrophoresis, PFGE) y tipificación de secuencias multilocus (multilocus sequence typing, MLST). La presencia de genes que codifican para BLEE se determinó mediante el uso de PCR y secuenciación. La prueba de sensibilidad a los antimicrobianos se realizó por microdilución del medio.
ResultadosEl análisis de PFGE reveló un alto grado de diversidad genética en el grupo de sensibles y de MDR no productores de BLEE. Sin embargo, la población de E. coli MDR productora de BLEE era menos diversa y se detectó un grupo grande de aislados formado por productores de CTX-M-15 y ST131.
ConclusionesEl presente estudio reveló que la población de E. coli MDR productora de BLEE era menos diversa que el grupo MDR no productor de BLEE, y que ST131 era dominante en los aislados de productores de CTX-M-15, lo que refleja la propagación de esta exitosa línea de MDR.
Urinary tract infections (UTIs) are highly prevalent worldwide and represent a clinical problem in both, community and hospital settings. Empirical therapy has been effective for years because susceptibility profiles among Gram-negative uropathogens have been relatively stable over the time. However, the emergence of multidrug resistance (MDR) has radically changed the situation with the emergence and dissemination of strains harboring extended-spectrum β-lactamases (ESBL) and less frequently class C β-lactamases and carbapenemases.1 MDR is often associated with worldwide-disseminated high-risk clones such as E. coli sequence type (ST) 131, which is considered a high-risk E. c clone mostly associated with UTI and bacteraemia.2
The dissemination of ESBL-producing E. coli has been extensively reported,3 however only a few studies, in different countries and human populations, have specifically studied the heterogeneity in population structure among susceptible, resistant and resistant by ESBL producing E. coli.4,5
This study aims to compare the population structure of three different representative groups of E. coli isolates causing UTI and also to evaluate alternative antimicrobial agents in each MDR group.
Material and methodsProspective collection of MDR and susceptible E. coli isolates.Strains of E. coli isolated from patients with urinary tract infection were collected between March and April 2017 in order to form three different groups: non ESBL-MDR, ESBL-MDR and a group susceptible to all the antibiotics. The samples were collected from 6 different medical areas representative of Madrid and responsible for the care of around 1,460,000 inhabitants.
In this study, MDR was defined as resistance at least to amoxicillin, ciprofloxacin, gentamicin and trimethoprim sulfamethoxazole according to their clinical MIC breakpoints.6 Suspected ESBL-producing isolates were phenotypically characterized using double-disk synergy.
Bacterial identification and antimicrobial susceptibility testingBacterial identification was performed by MALDI-TOF MS (Bruker Daltonik GmbH, Leipzig, Germany). Antibiotic susceptibility testing was performed by broth microdilution (MicroScan, Beckman Coulter, Inc; Brea, California) and interpreted according to the European Committee for Antimicrobial Susceptibility Testing (EUCAST) breakpoints.7
Susceptibility to ceftolozane/tazobactam was tested by E-test (Liofilchem, Roseto degli Abruzzi, Italy) at a fixed concentration of tazobactam of 4mg/L.
A chi square test (http://www.socscistatistics.com/tests/chisquare/) was applied to analyze differences in susceptibility to alternative antibiotics among non ESBL and ESBL groups. A p-value <0.05 was considered as statistically significant.
Molecular characterization of ESBLThe presence of genes encoding ESBL (blaSHV, blaCTX-M and blaTEM) was determined by using PCR and sequencing.8
Population structure analysisEpidemiological relatedness was studied by PFGE of XbaI-digested DNA.8
MLST was performed on randomly selected representative isolates from each clonal type (https://enterobase.warwick.ac.uk/species/ecoli/allele_st_search).
A Simple Diversity Index (SDI) was calculated in order to quantify the genetic variability in each group. The index was calculated as the total number of different PFGE profiles of each group/total number of strains in each group×100.
ResultsAntimicrobial susceptibility to alternative antibioticsAntimicrobial susceptibility results are summarized in Table 1. No statistically significant differences between both MDR groups were observed.
Antibiotic susceptibility of non-ESBL-MDR E. coli isolates and ESBL-MDR E. coli isolates.
| Antibiotics | Non-ESBL MDR | ESBL-MDR | p-value (<0.05) | ||
|---|---|---|---|---|---|
| MIC90* | Susceptibility (%) | MIC90* | Susceptibility (%) | ||
| Fosfomycin | ≤32 | 94.3 | >64 | 85.7 | 0.23 |
| Nitrofurantoin | ≤32 | 94.3 | 64 | 100 | NA |
| Piperacillin/tazobactam | 64 | 85.8 | 64 | 88.6 | 0.72 |
| Amoxicillin/clavulanate | >16 | 68.6 | 16 | 54.3 | 0.22 |
| Ceftolozane/tazobactam | ≤0.5 | 100 | 1 | 94.3 | NA |
| Ertapenem | ≤0.5 | 100 | ≤0.5 | 100 | NA |
| Imipenem | ≤1 | 100 | ≤1 | 100 | NA |
| Meropenem | ≤1 | 100 | ≤1 | 100 | NA |
| Amikacin | ≤8 | 100 | ≤8 | 100 | NA |
NA: not applicable.
Only 2 isolates from the ESBL MDR group were resistant to ceftolozane/tazobactam (MIC=2mg/L and MIC=3mg/L) while all the non-ESBL MDR strains were susceptible. A slight but non-significant increase in MIC was observed in the ESBL MDR group respect to the non-ESBL MDR group (MIC 50: 0.5mg/L and 0.38mg/L, respectively; MIC 90: 1mg/L and 0.5mg/L, respectively).
Molecular characterization of ESBL and molecular typingAll of the suspected ESBL-producing MDR strains were confirmed by PCR and sequencing. 71.4% produced CTX-M-15; 8.6% produced CTX-M-14; 8.6% produced SHV-12, 2.8% produced CTX-M-65; 2.8% produced CTX-M-27 and 5.7% co-produced both CTX-M-15 and CTX-M-14.
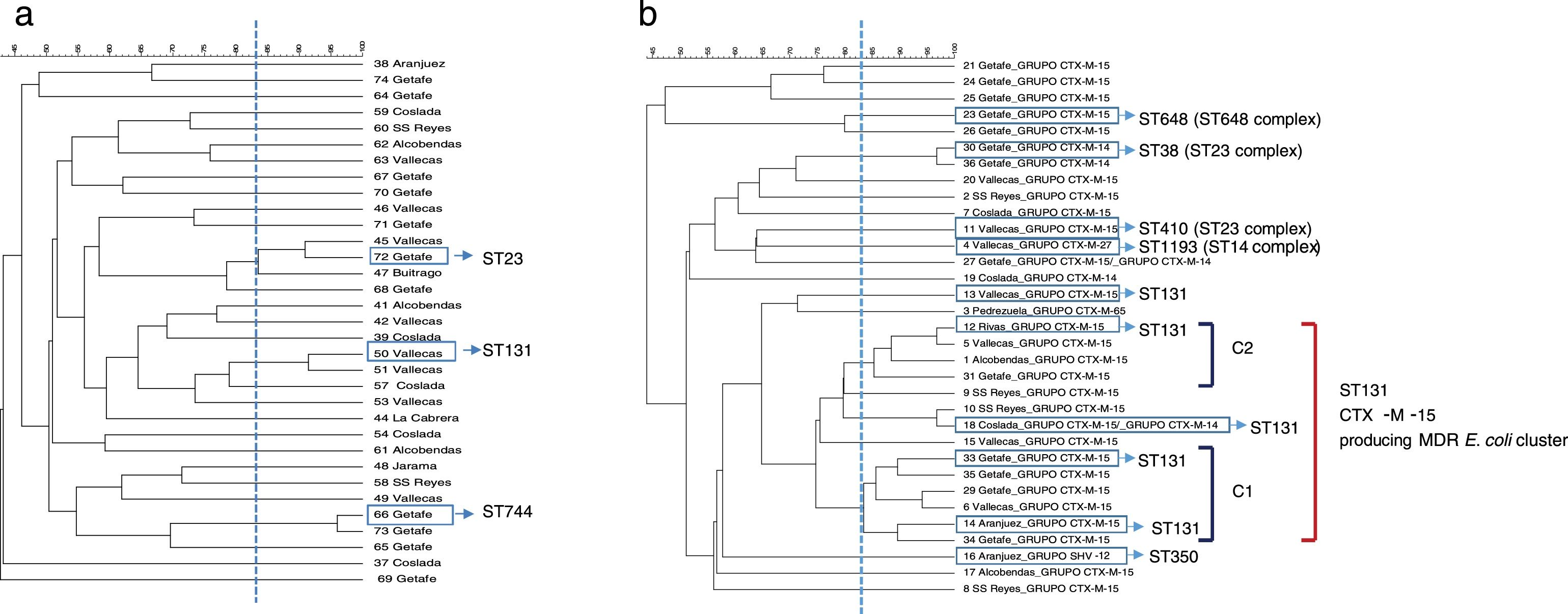
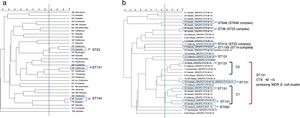
PFGE analysis revealed a high degree of genetic diversity in susceptible and non ESBL-MDR groups. Thirty-three strains from the non ESBL-MDR group were analyzed by PFGE and 30 different patterns were observed (SDI=90.9) (Fig. 1a). However, the ESBL-MDR E. coli population was less diverse and 24 different patterns were observed (SDI=72.7) respect to 33 strains analyzed (Fig. 1b). Two defined clusters: C1 (6 isolates) and C2 (4 isolates), consisting of CTX-M-15 producing isolates, were detected with a genetic linkage 83%. Representative isolates from each cluster were analyzed by MLST and all of them belonged to ST131 complex. The CTX-M-14 producing isolates belonged to ST23 complex. No PFGE patterns were obtained from two of the three SHV-12 producing isolates detected, but 2 of them studied by MLST belonged to CC350. The CTX-M-27 producing isolate belonged to ST1193 (ST14 complex). Three PFGE clusters were detected in the non ESBL-MDR group (Fig. 1a); by MLST these clusters belonged to ST131, ST744 and ST23. All the isolates from the susceptible group were different (SDI=100) and no clonal association was detected (data not shown).
All the ESBL-MDR E. coli isolates resistant to piperacillin/tazobactam (11.4%), fosfomycin (14.3%) and ceftolozane/tazobactam (5.7%) produced CTX-M-15.
Eighty percent of fosfomycin resistant isolates and only 25% of piperacillin/tazobactam resistant isolates grouped in the ST131-CTX-M-15-producing-MDR E. coli cluster (C1 and C2). The two isolates resistant to ceftolozane/tazobactam were not clonally related.
DiscussionMDR in urine isolates is often associated with the presence of ESBL genes, as well as aminoglycoside and quinolone resistance.9 Previous studies have reported that the emergence and spread of different high-risk clones, such as E. coli ST131 or K. pneumoniae ST258, have been largely responsible for the sudden increase in MDR.10 Specifically, a recent study in Spain has addressed the prevalence and dynamics of E. coli ST131 among contacts of infected patients.11 As a starting point, our group reported a recent local retrospective study in which it was analyzed the evolution of multidrug resistance (using the same MDR markers) in urine samples from both, outpatients and inpatients, over a 12-year period. This study showed that MDR is a phenomenon that is increasing not only in hospitalized, but also at a community level.6 Following with the same idea, the present study was designed to take a new step in this direction and describe the diversity in population structure between susceptible, MDR (non-ESBL) and MDR-ESBL producing E. coli irrespective of the origin (community acquired- or hospital acquired UTI). The PFGE analysis showed that there was a great clonal diversity among isolates in non ESBL-MDR group with smaller clusters while the ESBL-MDR group was rather less diverse and more clonal. Almost half of the ESBL-MDR isolates (14 isolates, 40%) were grouped in a large cluster and all of them belonged to a ST131 complex (inferred by the MLST results obtained) and produced CTX-M-15 beta-lactamase. The MDR strains selected in this study are representative of a significant human population in a country with a high consumption of antibiotics,12 and corroborate previous studies in which the ESBL population in non-hospitalized patients was less diverse and was dominated mostly by ST131.4 Other studies have reported similar results in samples from a different origin such as bloodstream infections 5,13 or according the patient characteristics.14
The predominance of the MDR ST131 clone as a cause of UTIs and bacteraemia represents a challenge for the treatment of these infections. Recent studies have demonstrated that β-lactam/β-lactamase-inhibitor combinations are suitable alternatives to carbapenems for treating patients with UTI or bloodstream infections due to ESBL-producing E. coli.15,16 We found a high rate of in vitro susceptibility to piperacillin/tazobactam among MDR ESBL-producing isolates (88.6%). Only four isolates were resistant to piperacillin/tazobactam among MDR ESBL-producing isolates, three of which were susceptible to ceftolozane/tazobactam. In total, 33 of 35 MDR ESBL-producing isolates (94.3%) were susceptible to ceftolozane/tazobactam. These findings are in line with previous studies carried out at different countries in which ceftolozane/tazobactam has demonstrated excellent overall in vitro activity against the vast majority of E. coli isolates including ESBL-phenotype and MDR strains.17
It should be also taken into account the percentage of susceptibility to antibiotics considered as the first line for empirical treatment of acute uncomplicated cystitis such as fosfomycin and nitrofurantoin. We found higher rates of resistance, but not significant, to fosfomycin among MDR ESBL-producing isolates (14.3%) with respect to the MDR non-ESBL isolates (5.7%), result that is in accordance with other studies in which the presence of ESBL entails a higher resistance to fosfomycin.18
The strength of the report is that the study was carried out in urine samples taken from 6 different medical areas which are responsible for the care of around 1,460,000 inhabitants (25 per cent of the Autonomous Community of Madrid population) and therefore, representative of a large area of Madrid. However, the study has also some limitations; the relatively small number of isolates characterized and the lack of detailed information about the patient characteristics.
Overall, these findings suggest that ST131 dominate the ESBL-producing lineages in the MDR uropathogenic E. coli in a representative area of Madrid. Therefore, molecular epidemiology and clonal group surveillance studies are necessary to evaluate the evolution over the time of MDR high-risk clones due to their capability to disseminate and to acquire more antimicrobial resistant determinants.
Ethical approvalNot required.
FundingD.M.A has received a research grant from the Investigator Initiated Studies Program economically supported by Merck Sharp and Dohme (MSD).
This work was also supported by Plan Nacional de I+D+i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16CIII/0004/0002) – co-financed by European Development Regional Fund ERDF “A way to achieve Europe”, Operative program Intelligent Growth 2014–2020.
Conflict of interestThe authors declare that they have no conflict of interest.