The presence of colonised patients is one of the main routes for the spread of multiresistant bacteria, and its containment is a clinical and public health priority. Surveillance studies are essential for early detection of colonisation by these bacteria. This article discusses the different microbiological methods, both based on culturing and molecular methods, for detection of carriers of multiresistant bacteria. Those species with a high clinical/epidemiological impact or generating therapeutic difficulties are included: Methicillin-resistant Staphylococcus aureus, Enterococcus spp. resistant to glycopeptides, enterobacteriaceae producing extended spectrum β-lactamases and plasmid-mediated AmpC, carbapenemases producing enterobacteriaceae, Acinetobacter baumannii and multiresistant Pseudomonas aeruginosa. The information in this document should be considered as a structure matrix to be tailored to the specific needs of each centre.
La existencia de pacientes colonizados es una de las principales vías de propagación de las bacterias multirresistentes, y su contención una prioridad asistencial y de salud pública. Los estudios de vigilancia son imprescindibles para una detección precoz de la colonización por estas bacterias. Este artículo aborda los diferentes métodos microbiológicos, basados en el cultivo y moleculares, para la detección del estado de portador de bacterias multirresistentes. Se incluyen aquellas especies de mayor interés debido a su impacto clínico/epidemiológico y a las dificultades terapéuticas que generan: Staphylococcus aureus resistente a meticilina, Enterococcus spp. resistente a los glucopéptidos, enterobacterias productoras de β-lactamasas de espectro extendido o β-lactamasas plasmídicas de tipo AmpC, enterobacterias productoras de carbapenemasas, Acinetobacter baumannii multirresistente y Pseudomonas aeruginosa multirresistente. La información recogida en este documento debe considerarse como una estructura matriz que deberá adaptarse a las necesidades específicas de cada centro.
Resistance of some of the major bacterial pathogens to multiple antibiotics is increasing,1–3 thereby limiting treatment options. Multidrug resistance (MDR), defined as non-susceptibility to at least one antibiotic in three or more families,4 can affect Gram-positive and Gram-negative bacteria. However, of particular importance is the spread of Gram-negative bacteria which are extensively drug-resistant (XDR: non-susceptible to at least one antibiotic in all families except one or two) and pandrug-resistant (PDR: non-susceptible to all antibiotics in all families normally used in the treatment of the bacteria in question),4 including Acinetobacter baumannii (A. baumannii), Pseudomonas aeruginosa (P. aeruginosa) and some species of Enterobacter. MDR bacteria usually have a high capacity for epidemic spread, not only in-hospital but also inter-hospital and out-of-hospital.1,5 Colonised patients represent one of the main routes for the propagation of these bacteria,6,7 and their containment has been recognised as a clinical and public health priority by the principal national and international institutions. Local microbiological surveillance studies are needed to enable early detection of patients colonised by this type of bacteria. EUCAST has guidelines for the detection of resistance mechanisms of clinical and epidemiological importance.8
The Spanish Society of Infectious Diseases and Clinical Microbiology (Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica, SEIMC) recently published a clinical microbiology procedure entitled “Microbiological methods for surveillance of carrier status of multidrug-resistant bacteria”.9 The purpose of this article is to summarise the main points of the procedure. We have covered the most relevant pathogenic species in terms of their clinical and epidemiological impact: Methicillin-resistant Staphylococcus aureus (MRSA); glycopeptide-resistant Enterococcus spp. (GRE); Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) or plasmid-mediated AmpC β-lactamases (pAmpC); carbapenemase-producing Enterobacteriaceae (CPE); multidrug-resistant A. baumannii; and multidrug-resistant P. aeruginosa. The general information presented in this document should be considered as a matrix structure that will benefit from suitable adaptation to the specific needs established by the multidisciplinary nosocomial infection control team at each centre.
Clinical and epidemiological impactMethicillin-resistant Staphylococcus aureus (MRSA)Apart from routinely colonising humans, the microorganism S. aureus can also behave as an opportunistic pathogen and be the cause of a wide range of infections. In 2013, data from the European Antimicrobial Resistance Surveillance Network (EARS-Net) showed the prevalence of MRSA in bacteraemia to be 22.6% in Spain and 18% in the participating European countries overall. Within Spain, the prevalence varies considerably depending on the geographical area and type of hospital. The clinical and epidemiological implications make it necessary to establish adequate detection systems.10 The main mechanism of resistance to methicillin is the production of a penicillin-binding protein (PBP2a) encoded by the mecA gene, which has low affinity for β-lactam antibiotics, with the exception of a new class of cephalosporins such as ceftaroline or ceftobiprole. In 2011, a homologous gene named mecC was detected.11 MRSA-mecC strains have been isolated in many countries, including Spain, from humans and a wide variety of animals. The detection of the mec gene made it possible to identify other rare strains of S. aureus with low levels of resistance to methicillin as a result both of changes in the PBP (moderately resistant S. aureus [MODSA]) and overproduction of β-lactamase (borderline oxacillin-resistant S. aureus [BORSA]). The protocol described in this procedure refers to S. aureus containing the mec gene.
Glycopeptide-resistant Enterococcus spp. (GRE)Enterococcus faecalis (∼80 to 90%) and Enterococcus faecium (∼5 to 10%) are the species that cause the majority of infections in humans. These microorganisms are responsible for approximately 10% of healthcare-related infections. Also important is their intrinsic resistance to multiple antibiotics, and their ability to develop resistance to other antibiotics, either by acquisition of genes located in plasmids or transposons (e.g. glycopeptide resistance) or by chromosomal mutations (e.g. fluoroquinolone resistance). In the US, there was a rapid increase in nosocomial outbreaks of GRE that led to an endemic situation in certain hospitals. In Europe, however, GRE was initially isolated from community sources (healthy people and animals) because of widespread use of avoparcin, a glycopeptide used as a growth promoter for farm animals until it was banned in 1997. Data from the EARS-Net in 2013 showed significant differences in the prevalence of GRE in different European countries. In Spain, the prevalence of vancomycin-resistant E. faecium was 0.9% in 2013. Despite the low percentage, there are relatively frequent reports of hospital outbreaks.
The acquired resistance to glycopeptides by enterococci is mediated by nine different “van” operons (vanA, vanB, vanC, vanD, vanE, vanG, vanL, vanM, vanN). Of these, vanA and vanB are by far the most common in E. faecalis and E. faecium. The vanA gene confers inducible high-level resistance to vancomycin and teicoplanin, while the vanB gene confers inducible high or moderate resistance to vancomycin and remains susceptible to teicoplanin. The species of E. faecium or E. faecalis with resistance to vancomycin (MIC>4mg/l) are those of most epidemiological importance and they require microbiological surveillance.12
Enterobacteriaceae producing extended-spectrum β-lactamases, plasmid-mediated AmpC β-lactamases and carbapenemasesThe spread of Enterobacteriaceae resistant to broad-spectrum cephalosporins, mainly ESBL, has become one of the main threats to antibiotic treatment of infections caused by these bacteria. In Spain, ESBL-producing E. coli and K. pneumoniae, especially CTX-M-15 and SHV-12, have spread rapidly in recent years and now have an overall prevalence of around 15%. Although less common, pAmpC is emerging as an additional threat to the activity of broad-spectrum cephalosporins. The major types of pAmpC are CIT and DHA, which mainly affect E. coli, K. pneumoniae and Proteus mirabilis. The frequent association of ESBL and pAmpC production with resistance to other non-β-lactam antibiotics has led to an increase in the use of carbapenem antibiotics and the development of strains resistant to them. At present, one of the major threats in the area of antibiotic resistance is the rapid spread of MDR and XDR carbapenemase-producing strains of Enterobacteriaceae (CPE). These enzymes are able to hydrolyse almost all β-lactam antibiotics, including carbapenems. The majority are grouped according to the Ambler molecular classification as follows: (1) class A, mainly KPC enzymes; (2) class B or metallo-β-lactamases (MBLs), mainly VIM, IMP and NDM enzymes; and (3) class D, mainly OXA-48. A multi-centre study conducted in 2013 (GEIH-GEMARA-REIPI project) showed that OXA-48 (71.5%) and VIM-1 (25.3%) were the most common carbapenemases in Enterobacteriaceae in Spain and K. pneumoniae (74.4%) the most affected species.1 Also detected was extensive spread of a number of high-risk clones of K. pneumoniae, such as ST11/OXA-48, ST15/OXA-48, ST405/OXA-48 and ST11/VIM-1.1 Although less common, other carbapenemases, such as KPC and NDM, are also on the increase in Spain and causing significant in-hospital and inter-hospital outbreaks.13 At the end of 2015, the scenario in Spain was of an inter-regional spread of CPE.13 There are very few therapeutic alternatives in these cases and they are almost never optimal.
While surveillance of colonisation by ESBL- and carbapenemase-producing Enterobacteriaceae should cover all species, surveillance of pAmpC should concentrate on the species that do not have chromosomal AmpC and have demonstrated their ability to spread these enzymes; primarily K. pneumoniae and P. mirabilis.
Multidrug-resistant Acinetobacter baumanniiA. baumannii is an opportunistic pathogen involved in infections such as mechanical ventilator-associated pneumonia, endocarditis, skin and soft tissue infections, meningitis and urinary tract infections, especially among patients admitted to ICUs.
The genus Acinetobacter consists of 30 species with an associated name and nine genomic species defined through DNA-DNA hybridisation studies. A. baumannii shows an extraordinary ability to acquire resistance to multiple antibiotics. The most recent national multicentre study (GEIH-GEMARA-REIPI-Ab2010 project), which included 446 isolates of A. baumannii obtained from 43 Spanish hospitals,3 detected a high prevalence of reduced susceptibility to most of the antimicrobial agents: >94% (ceftazidime, piperacillin and ciprofloxacin); 82–86% (carbapenems and tetracycline); and 60–70% (tobramycin, sulbactam, gentamicin and doxycycline). Compared to the data from a previous study in 2010,14 these isolates were more resistant to ceftazidime, carbapenems, doxycycline, sulbactam and colistin; the majority were MDR or XDR. The same study revealed that the incidence rate of colonisation or infection by A. baumannii increased significantly from 0.14 in 2000 to 0.52 in 2010 (p<0.001). In addition, the epidemiological study using MLST (multilocus sequence typing) detected an increase in the clonal ST2 group in 2010 which showed greater resistance to imipenem and was associated with an increased risk of sepsis.6
Different types of β-lactamases have been described in A. baumannii, but the acquired carbapenemases in class D (OXA-23, -24, -58, -143 and -235) have the greatest clinical and epidemiological impact. Reduction in permeability, overexpression of active efflux systems, production of enzymes capable of modifying aminoglycosides and chloramphenicol, methylases, and modifications of the target interaction site are also implicated in A. baumannii resistance to antibiotics.
The epidemiological factors involved in hospital-acquired Acinetobacter spp. are well known. In hospital outbreaks, the spread is aided by their ability to survive on dry surfaces, to be transmitted from patient to patient on the hands of staff and to contaminate the hospital environment. In Spain, outbreaks of A. baumannii carrying OXA-23, OXA-24 and OXA-58 carbapenemases have been reported.
Multidrug-resistant Pseudomonas aeruginosaP. aeruginosa is one of the most common causes of nosocomial infection and chronic respiratory infection in patients with cystic fibrosis. The prevalence of MDR strains exceeds 30% worldwide, and this includes Spanish hospitals; approximately half of the MDR strains may also be XDR.2 This increasing prevalence is due to the extraordinary ability of P. aeruginosa to select chromosomal mutations that generate resistance and the increasing production of exogenous resistance determinants.15
The main mechanism of resistance to penicillins or cephalosporins is the selection of mutants with constitutive overproduction (derepression) of the inducible chromosomal cephalosporinase AmpC.16 Another of the mechanisms of mutational resistance is the inactivation of the porin OprD, which confers resistance to imipenem and decreased susceptibility to meropenem. Lastly, overexpression of one of the multiple efflux pumps, mainly MexAB-OprM and MexXY-OprM, contributes significantly to the resistance phenotypes.16 Although proportionally much less common than mutational resistance, detection of transferable genetic elements carrying carbapenem or ESBL genes is increasingly common. Recent studies show that most of the strains producing carbapenemases or ESBL belong to so-called high-risk clones, mainly ST235, ST111 and ST175.2,5 Particularly relevant are the MBLs, whose prevalence in Spain has increased more than 10-fold (from 0.08 to 1%) in five years (2003–2008), and which have been responsible for significant epidemic outbreaks.16 The MBLs described in P. aeruginosa include VIM, IMP, SPM, GIM, SIM, NDM, AIM and FIM.17 By far the most common MBL found in Spain is VIM-2. Important among the class A carbapenemases are the GES and among ESBLs, the OXA.18
Collection, transport, storage and processing of the sampleTaking, transporting and storing samples for surveillance cultures should be carried out following the general recommendations of SEIMC Clinical Microbiology Procedure No.° 1a, “Collection, transport and general processing of samples in the Microbiology laboratory”.19 It should be explicitly stated that the request corresponds to a “culture for microbiological surveillance”. The bacteria and resistance problem to be monitored should also be stated as this information is needed to determine the adequacy of the type of sample for the requested test (Table 1). The sample should be processed as quickly as possible and within 24h; if it is not to be processed immediately, it should be stored at 2–8°C to facilitate the recovery of the bacteria being tested for and to prevent overgrowth of commensal microbiota.
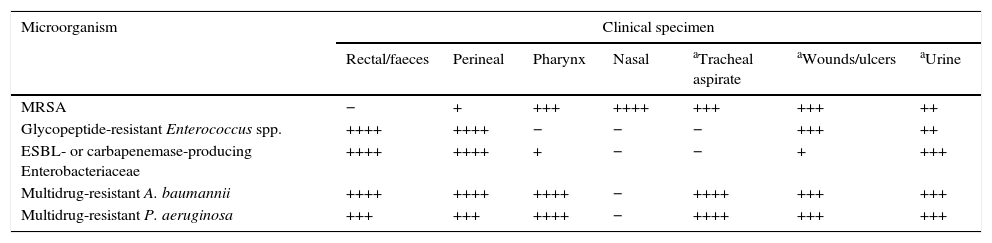
Guidance on the qualitative interest of different clinical specimens for the research of multidrug-resistant pathogens for epidemiological purposes.
| Microorganism | Clinical specimen | ||||||
|---|---|---|---|---|---|---|---|
| Rectal/faeces | Perineal | Pharynx | Nasal | aTracheal aspirate | aWounds/ulcers | aUrine | |
| MRSA | − | + | +++ | ++++ | +++ | +++ | ++ |
| Glycopeptide-resistant Enterococcus spp. | ++++ | ++++ | − | − | − | +++ | ++ |
| ESBL- or carbapenemase-producing Enterobacteriaceae | ++++ | ++++ | + | − | − | + | +++ |
| Multidrug-resistant A. baumannii | ++++ | ++++ | ++++ | − | ++++ | +++ | +++ |
| Multidrug-resistant P. aeruginosa | +++ | +++ | ++++ | − | ++++ | +++ | +++ |
In previous studies of MRSA colonisation, nasal exudate is the most suitable single sample, although double (nasal-pharyngeal or nasal-perineal) and especially triple (nasal-pharyngeal-perineal) sampling has greater susceptibility. The gastrointestinal tract is the main reservoir of multidrug-resistant Enterobacteriaceae and GRE. The most usual samples for surveillance cultures of GRE are therefore rectal smears and faeces. In circumstances where an environmental contamination study is required, samples from surfaces and medical instruments in contact with the patient should be analysed. Given the complexity of the epidemiology of multidrug-resistant A. baumannii infections, it is appropriate to consider sampling both the patient and the environment. The most commonly obtained surveillance samples include sputum and tracheostomy exudate, and wound, axilla/groin and rectal smears. Screening effectiveness is increased by sampling different anatomical areas of the body. Surveillance cultures for multidrug-resistant P. aeruginosa also include sampling of patients, the environment and the healthcare teams. Respiratory samples such as sputum, pharyngeal swabs, tracheostomy exudate, etc. are particularly useful. Intestinal colonisation by multidrug-resistant P. aeruginosa is detected relatively frequently in rectal smears.
Culture-based methodsMethicillin-resistant Staphylococcus aureusSelection of culture media and incubation conditionsDetection of colonisation by MRSA is essentially based on culture in selective and differential media. The combination of a mannitol salt agar plate (MSA or Chapman medium) and an MSA-cefoxitin plate (4mg/l) allows the detection of MRSA and possibly also colonisation by methicillin-susceptible S. aureus (MSSA). Some authors have estimated sensitivity of 96% and specificity of 100% after 48h of incubation.20 A recent study evaluating five chromogenic media (Brilliance MRSA agar [Oxoid], ChromoID [bioMérieux], MRSASelect [Bio-Rad], CHROMagar [CHROMagar Microbiology], and BBL-CHROMagar [BD Diagnostics]), showed sensitivity of 90% for the Brilliance MRSA agar medium and 81–83% for the rest of the media, and specificity of 87% for Brilliance MRSA agar and 97–99% for the rest of the chromogenic media.21 Pre-incubation of the samples in a selective liquid enrichment medium increases the sensitivity of these media. According to published studies, there are no absolute recommendations for or against a particular chromogenic medium.
Confirmation tests and criteria for interpreting resultsIn view of possible false positive results, confirmation may be necessary in both identification and resistance. In the first case, a coagulase test, latex agglutination or MALDI-TOF identification are recommended. To confirm resistance, using a cefoxitin disc (30μg) and carrying out the reading at 24h (<22mm) (EUCAST 2015) is recommended. In automated systems which include cefoxitin and oxacillin, although these results usually correspond, in the event of discrepancies, interpretation of conflicting results should lean towards the resistant outcome (EUCAST 2015). The resistance profile “oxacillin-S and cefoxitin-R” may be indicative of mecC-positive MRSA. In such cases, the recommendation is to perform phenotypic and genotypic studies (mecA or mecC) of the S. aureus strains. The resistance can also be confirmed by the agglutination of latex particles coated with monoclonal antibodies of the PBP2a protein. Isolates that produce small amounts of this protein may give weak results or slow the agglutination reaction. Moreover, mecC-positive MRSA strains will not be detected by this assay.
Reporting of the resultsIf at 48h there are no colonies with the characteristic colouration it will be reported as, “Methicillin resistant S. aureus not isolated” (MRSA negative). If a microorganism with the characteristic colouration is isolated, it will be reported as, “Methicillin resistant S. aureus isolated” (MRSA positive).
Glycopeptide-resistant Enterococcus spp.Selection of culture media and incubation conditionsMost common are the selective and differential media that enable rapid detection of GRE from highly contaminated samples. These include Enterococcosel agar (BEAV) medium (BD Diagnostics) and several recently introduced chromogenic media, such as ChromoID VRE (bioMérieux), CHROMagar VRE (BD Diagnostics), VRESelect (Bio-Rad) and Spectra VRE (Remel). These media incorporate chromogens that facilitate the distinction between E. faecium and E. faecalis. Some studies show sensitivity ranging from 92% to 95%, and specificity from 96% to 99%.22 In general, the increase in incubation time to 48h slightly decreases specificity. The use of an enrichment medium prior to inoculation in the chromogenic medium may increase sensitivity. There are no absolute recommendations for or against a particular chromogenic medium.
Confirmation tests and criteria for interpreting resultsIn the BEAV agar medium, the appearance of small translucent colonies with black or brown pigmentation around them suggests the presence of GRE. If a chromogenic medium has been used, interpretation of the culture will depend on the colour of the colony according to the criteria of the manufacturer. As false positive results do occur (other species of enterococci, Streptococcus spp., some Gram-negative bacilli and Candida spp.) it may be necessary to confirm identification by biochemical testing or by MALDI-TOF.
Reporting of the resultsIf the culture is positive for GRE, it will be reported as “Vancomycin-resistant [E. faecium/E. faecalis] isolated”. If the culture is negative for GRE after 48h, it will be reported as “Glycopeptide-resistant enterococci not isolated”. Isolation of E. gallinarum and E. casseliflavus with low levels of intrinsic resistance to vancomycin (vanC) should not be reported when isolated in surveillance cultures, because their true epidemiological significance is unknown.
Enterobacteriaceae producing extended-spectrum β-lactamases, plasmid-mediated AmpC β-lactamases and carbapenemasesSelection of culture media and incubation conditionsAs a general rule, it is necessary to use selective media to recover Enterobacteriaceae resistant to third-generation cephalosporins or carbapenem antibiotics and prevent the growth of susceptible commensal microbiota. Media such as MacConkey agar or Drigalski agar supplemented with cefotaxime or ceftazidime have proved useful for screening for ESBL and pAmpC. In these media, Enterobacteriaceae can grow with clavulanic acid-inhibitory chromosomal β-lactamases which, on overexpression, can hydrolyse cephalosporins. These include the β-lactamase K1 from Klebsiella oxytoca, SHV-1 from K. pneumoniae and the CepA cephalosporinases from Proteus vulgaris and Proteus penneri. Different chromogenic media have been developed for the selective isolation of ESBL-producing Enterobacteriaceae, such as ChromID ESBL (bioMérieux), Brilliance ESBL agar (Oxoid) and CHROMagar™ ESBL.
Most of the selective media for detection of resistance to third-generation cephalosporins can be used for the screening of carbapenemases, as long as a subsequent check is performed. The drawback of this approach is the large amount of extra work involved in rechecking all the instances of ESBL isolation. Furthermore, with this approach, carbapenemase OXA-48-producing isolates not producing ESBL or AmpC would be lost.
Different selective media have been used for the specific detection of CPE, mainly MacConkey agar with low carbapenem concentrations (1mg/l imipenem) or with 10μg imipenem or ertapenem disks. The sensitivity of these techniques can be optimised with prior enrichment by introducing the rectal swab into a BHI broth with a 10μg imipenem disk. For any of these techniques, meropenem should be considered as the carbapenem antibiotic with the best balance between sensitivity and specificity for detecting CPE.
Commercial chromogenic media have been developed in recent years for the detection of CPE. These include chromID media (bioMérieux, France), including the CARBA medium, the OXA-48 medium and the CARBA SMART medium; the CRE Brilliance medium (Thermo Fisher Scientific, UK) and the SUPERCARBA medium, which contains Drigalski agar, ertapenem, cloxacillin and zinc sulfate. Apart from the first two, which need to be used together, these media have demonstrated a high sensitivity for detecting any type of carbapenemase.
Confirmation tests and criteria for interpreting resultsThe specificity of these media is not always as high as their sensitivity, so any growth must be confirmed. Once growth of Enterobacteriaceae has been documented, a phenotypic study should be performed to confirm the presence of an ESBL, a pAmpC or a carbapenemase.8,9 The production of carbapenemase must be tested in isolates with carbapenem MICs higher than the epidemiological cut-off points established by EUCAST.8,9 There are different tests for phenotypic confirmation of the production of carbapenemases, such as the modified Hodge test, the measurement of hydrolysis of carbapenem antibiotics by spectrophotometry, methods based on specific inhibition of the different classes of carbapenemases, colorimetric methods based on change in pH (Carba NP and BlueCarba), and the carbapenem inactivation method (CIM).
The spectrophotometry techniques detect the specific hydrolysis of β-lactam antibiotics.9 These are reference techniques, but they are difficult to implement in the majority of Clinical Microbiology laboratories.
The colorimetric methods are based on detection of the change in pH produced by hydrolysis of the carbapenem using a colorimetric scale. These methods are very sensitive and specific and all steps need to be carefully followed to obtain optimal performance.9
The modified Hodge test is a simple and widely used technique for the detection of carbapenemase activity. Drawbacks include false negatives (mainly with MBL, which can largely be avoided by adding zinc sulfate to the medium) and false positives (mainly strains with AmpC overproduction plus loss of porins, which can largely be resolved by adding cloxacillin or oxacillin to the medium). However, taking the results in conjunction with carbapenemase inhibitor-based techniques provides quick and easy-to-interpret information. The existence of certain specific carbapenemase inhibitors (phenylboronic acid for class A and EDTA and dipicolinic acid for class B) enables the phenotypic characterisation of these carbapenemases from the recovery of carbapenem antibiotic activity in the presence of these compounds.9
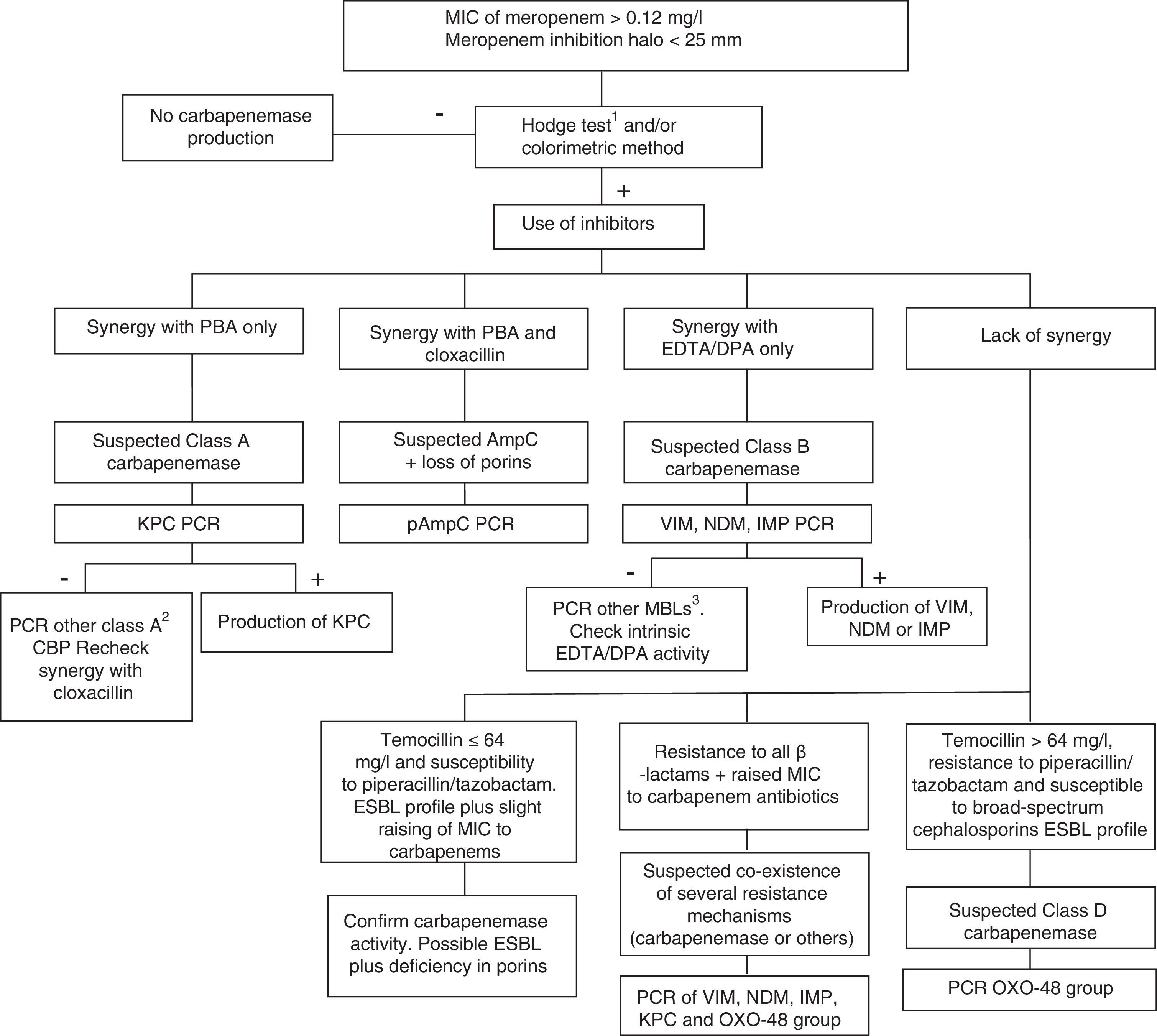
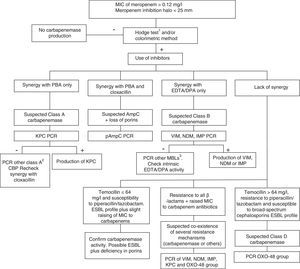
Class D carbapenemases, primarily OXA-48, hydrolyse carbapenem antibiotics without affecting (ceftazidime, cefepime) or only slightly affecting (cefotaxime) broad-spectrum cephalosporins. Nonetheless, OXA-48 producing strains have a high association, more in K. pneumoniae than in other species, with the production of ESBL. These enzymes do not have specific inhibitors, but they do have high-level resistance to temocillin and piperacillin/tazobactam (>64mg/l). However, resistance to temocillin and piperacillin/tazobactam are not specific OXA-48 markers, so for confirmation of class D carbapenemases, using a genotypic method or one of the recently marketed enzymatic methods is recommended. In general, the presence of mixed mechanisms of resistance to carbapenem antibiotics greatly hampers phenotypic techniques for identifying these mechanisms; molecular techniques are recommended for such cases. Fig. 1 shows an algorithm for the confirmation of mechanisms of resistance to carbapenem antibiotics in Enterobacteriaceae.
Algorithm for the confirmation of mechanisms of resistance to carbapenem antibiotics in Enterobacteriaceae. CBP: carbapenemase; DPA: dipicolinic acid; EDTA: ethylenediaminetetraacetic acid; ESBL: extended-spectrum β-lactamase; MBL: metallo-β-lactamases; PBA: phenylboronic acid. 1When using the Hodge test, it is recommended that it be performed and interpreted in conjunction with the techniques that use β-lactamase inhibitors. 2GES, SME, IMI, NMC. 3Among others, SIM, GIM, AIM, SPM.
Isolation of Enterobacteriaceae producing an ESBL, pAmpC or carbapenemase should be reported as, “[Species name] producing [resistance mechanism] isolated”. If the phenotypic results are very indicative of the class of carbapenemase produced, it should be reported as, “Carbapenemase-producing [species name] isolated, highly suspect of belonging to class [A or B or D]”. If after 48h the presence of Enterobacteriaceae producing these enzymes is not confirmed, it should be reported as, “Enterobacteriaceae producing [resistance mechanism] not isolated”.
Multidrug-resistant Acinetobacter baumanniiSelection of culture media and incubation conditionsA. baumannii grows very easily in conventional culture media. Resistant A. baumannii can be isolated using a conventional medium like MacConkey agar, supplemented or not with antibiotics such as gentamicin (8mg/l) or cefotaxime (2mg/l). Another culture medium which has been used for the selective isolation of Acinetobacter is LAM (Leeds Acinetobacter medium), which contains cefsulodin (15mg/l) and cefradine (50mg/l) to inhibit Gram-negative bacteria, and vancomycin (10mg/l) to inhibit Gram-positive bacteria. LAM also contains fructose and sucrose, sugars which are not fermented by the Acinetobacter species, leading to the colonies growing with a pink colouring. A Spanish study with LAM revealed a high positive predictive value for detecting A. baumannii (90.7%) from environmental samples. K. pneumoniae also grows in LAM, but it is easily distinguishable from A. baumannii.23
There are also other chromogenic media, such as CHROMagar Acinetobacter. This medium inhibits the growth of most Gram-positive cocci and yeast and uses an identification method that makes the Acinetobacter colonies grow red. With the addition of specific supplements, CHROMagar Acinetobacter has shown good sensitivity and specificity for the detection of carbapenem-resistant A. baumannii-A. calcoaceticus isolates, although further studies are required to achieve clinical validation. Whichever method used, the culture media should be incubated for 48h aerobically at 35–37°C.
Confirmation tests and criteria for interpreting resultsThe cultures are read after 24h incubation and, if negative, at 48h. Growth on MacConkey agar of gram-negative, oxidase-negative, catalase-positive diplococci (coccobacilli) suggests the presence of Acinetobacter spp. In view of the lack of specificity of the phenotypic tests, if A. baumannii is identified, it is advisable to refer to A. baumannii-A. calcoaceticus complex identification, because of the phenotypic similarity with other species such as A. pitii, A. nosocomialis and A. calcoaceticus. Molecular identification provides accurate identification of the species, but the complexity of the technique and the time required mean that routine application in Microbiology laboratories is difficult. Methods based on proteomics have enhanced species identification in the Acinetobacter genus.
Once the species of Acinetobacter is identified or it has been allocated to the genus Acinetobacter spp., the antibiogram is performed for classification as MDR, XDR, or PDR.4 However, in view of the complexity of resistance mechanisms, from a practical point of view in the case of Acinetobacter spp., the MDR nature will be defined by carbapenem resistance.
Reporting of the resultsIsolation and identification of multidrug-resistant Acinetobacter spp. (or A. baumannii or other species) will be reported as, “Multidrug-resistant Acinetobacter spp. isolated”. If after incubating for 48h the microorganism is not isolated, it will be reported as, “Acinetobacter spp. not isolated”. If within the incubation period a strain is isolated which does not show the multidrug-resistant profile, it will be reported as “Multidrug-resistant Acinetobacter spp. not isolated”.
Multidrug-resistant Pseudomonas aeruginosaSelection of culture media and incubation conditionsClinical strains of P. aeruginosa are resistant to the concentrations of cefotaxime (1–2mg/l) normally used in the selective media for microbiological surveillance of multidrug-resistant Enterobacteriaceae, so this may be used to detect P. aeruginosa colonisation. The culture media should be incubated aerobically at 35°C for 48h. In order to isolate MDR/XDR P. aeruginosa suitable selective media should be used, with MacConkey agar generally being used as a base. Two possible scenarios should be considered for the design: generic surveillance of MDR/XDR P. aeruginosa; or monitoring of a particular MDR/XDR strain in the context of an outbreak or endemic situation. In the first case, we need to choose a specific antipseudomonal which is the most representative possible of the P. aeruginosa multidrug-resistance problem. The most widely recommended is meropenem (concentration 1–2mg/l). For the monitoring of specific MDR/XDR strains, the most appropriate antibiotic(s) should be selected according to the phenotype. Lastly, the detection of strains producing carbapenemases or ESBL may be enhanced by the addition of cloxacillin, a potent AmpC inhibitor, to the selective medium.
Confirmation tests and criteria for interpreting resultsOnce the growth of P. aeruginosa has been documented, an antibiogram is necessary to define the MDR, XDR or PDR profiles.4 Evaluation of ceftazidime-avibactam or ceftolozane-tazobactam might also be useful for inference of the presence of acquired β-lactamases. Similarly, the inhibition of β-lactam resistance by cloxacillin is a marker of chromosome resistance (OprD+AmpC).24 Once the presence of an acquired β-lactamase is detected, phenotypic assays for ESBL or carbapenemases may be performed, including EDTA inhibition assays for MBL, modified Hodge test, or any of the available colorimetric methods for the detection of carbapenemase activity.18
Reporting of the resultsAt the very least it should be reported whether the sample is positive or negative for MDR P. aeruginosa. Ideally, the analysis should go further and differentiate, within the MDRs, the strains that have XDR profiles. Additionally, depending on the resources available at each centre, strains producing ESBLs or carbapenemases/MBLs should be reported in a specific manner.
Molecular methodsMolecular methods allow the rapid identification of a resistant microorganism by detecting the antibiotic resistance gene involved. The main advantage is that a result can be obtained on the same working day, around 1–3h after receipt of the sample. Worth highlighting are the greater sensitivity of molecular methods compared to culture-based techniques and the potential for direct application on clinical specimens, which is very important for making infection-control decisions. Drawbacks include the fact that only those genes designed in the test are identified and that they cannot completely replace microbiological culture, which enables identification of the species and the viability of the strain, and makes it possible to carry out antibiotic susceptibility or molecular typing tests.
The molecular methods most used in clinical practice and the most cost-effective are those based on real-time PCR with TaqMan probes, although there are other different or more complex modalities.
The higher financial cost of molecular testing with respect to culture-based methods is an important aspect when considering whether or not to introduce it in clinical practice. Generalisation of the use of molecular testing for the surveillance of multi-drug resistant bacteria is a subject for debate but, depending on the specific situation of each hospital, the type of patients and the epidemiological circumstances, it can be very useful and cost-effective.
Molecular methods for the detection of methicillin-resistant Staphylococcus aureus and glycopeptide-resistant Enterococcus spp.The commercial methods for the detection of MRSA approved for diagnostic use have been validated primarily for nasal samples (Table 2).21,25 The detection of GRE is performed mainly in samples of rectal/perianal swabs.
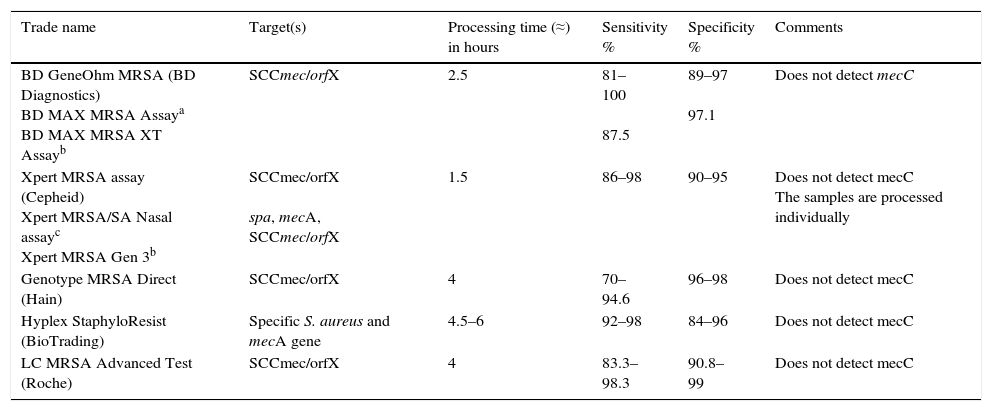
Characteristics of commercial molecular methods for detection of MRSA in nasal samples.
| Trade name | Target(s) | Processing time (≈) in hours | Sensitivity % | Specificity % | Comments |
|---|---|---|---|---|---|
| BD GeneOhm MRSA (BD Diagnostics) BD MAX MRSA Assaya BD MAX MRSA XT Assayb | SCCmec/orfX | 2.5 | 81–100 87.5 | 89–97 97.1 | Does not detect mecC |
| Xpert MRSA assay (Cepheid) Xpert MRSA/SA Nasal assayc Xpert MRSA Gen 3b | SCCmec/orfX spa, mecA, SCCmec/orfX | 1.5 | 86–98 | 90–95 | Does not detect mecC The samples are processed individually |
| Genotype MRSA Direct (Hain) | SCCmec/orfX | 4 | 70–94.6 | 96–98 | Does not detect mecC |
| Hyplex StaphyloResist (BioTrading) | Specific S. aureus and mecA gene | 4.5–6 | 92–98 | 84–96 | Does not detect mecC |
| LC MRSA Advanced Test (Roche) | SCCmec/orfX | 4 | 83.3–98.3 | 90.8–99 | Does not detect mecC |
Molecular tests for detection of MRSA should distinguish between MRSA colonisation and mixed colonisation by MSSA and methicillin-resistant coagulase-negative staphylococci. Some molecular tests combine the detection of a specific gene for S. aureus and the mecA gene, and others target the SCCmec/orfX binding region. Although this region has been considered MRSA-specific, it does present some variability, and that could cause reduced sensitivity in certain methods. Moreover, some strains of MSSA may contain fragments of the structure of the remaining SSCmec (mec-negative gene) and give false positives. Added to that, most commercial molecular assays do not detect the mecC gene. In general, these methods have high sensitivity and a high negative predictive value. In places with low prevalence, negative results are useful for ruling out MRSA, whereas positive results require confirmation by culture. In places with high prevalence, these methods could be used as a definitive test for identifying MRSA carriers.
The commercial methods which have been developed for the detection of GRE (BD GeneOhm VanR [BD Diagnostics, FDA approved], LC vanA/vanB detection assay [Roche Diagnostics, use in research], and Xpert vanA/vanB [Cepheid, use in research]) have vanA and vanB genes as molecular targets. These methods show limited sensitivity and, in particular, low specificity owing to the presence of the vanB gene in other species of bacteria. Performing these direct-sample tests for rapid detection of vanA and vanB genes does not exclude the need for the GRE surveillance culture.
The detection of MRSA DNA in clinical specimens will be reported as, “MRSA positive: MRSA DNA detected”; this is interpreted as MRSA nasal colonisation. If it is not detected, it will be reported as, “MRSA negative: MRSA DNA not detected”; MRSA nasal colonisation is ruled out.
In the case of molecular assays for the detection of GRE, the results are reported as follows: “VanA gene/vanB gene/or both detected. Culture required to confirm the results”; or “VanA/vanB genes not detected. Culture required to confirm results”.
Molecular methods for detecting extended-spectrum β-lactamases, plasmid-mediated AmpC β-lactamases and carbapenemases in Gram-negative bacilliThere are different molecular methods that enable the detection of acquired broad-spectrum β-lactam resistance genes in Gram-negative bacilli. They are classified according to the desired number of genes (single PCR or multiplex PCR) and the technique used (real-time PCR, microarray and pyrosequencing). DNA amplification by single PCR allows the identification of a single gene and requires specific primers for the desired target. Although it is possible to distinguish between very similar allelic variants with the design of primers that include the sites of variation between alleles, in general, for the characterisation of specific allelic variants a subsequent amplicon sequencing or the use of pyrosequencing is required. By using a single PCR with degenerate primers different variants of the same family can be detected.
The use of multiplex PCR allows the characterisation of different mechanisms in a single amplification reaction. Over the last ten years, several commercial multiplex PCR kits have been proposed for the detection of genes encoding pAmpC, ESBL and carbapenemases. Limitations of these techniques include the non-discriminative size of the amplicons, the use of different amplification conditions, cross-amplification with chromosomal β-lactamase genes and, in general, the inclusion of a limited number of genes.
Microarray is a very useful method for analysing a large number of genes in the same assay and detecting nucleotide variations of an allele. It is a versatile technology which is easy to apply and update, and which has demonstrated high sensitivity and specificity.
Loop-mediated isothermal amplification (LAMP) can be applied for the independent molecular detection of ESBL and carbapenemase genes.
In the case of P. aeruginosa, the main limitation for the detection of MDR by molecular techniques is that it is frequently mediated by chromosomal mutations. However, they are useful for the detection of transferable resistance mechanisms. There are many techniques available, many of them commercial, but they are generally only optimised for Enterobacteriaceae, which may affect the efficiency of the technique and mean that the choice of selected genetic targets is not the most appropriate for P. aeruginosa. This affects the ESBLs in particular, as the most common ESBLs in P. aeruginosa (OXA, PER, GES, VEB, etc.) are not usually included in the protocols designed for Enterobacteriaceae.
In the case of A. baumannii, the presence of genes encoding carbapenemases of the OXA type (families OXA-23, -24, -58, -143, -235 and ISAba1-OXA-51) is considered to be the main molecular marker of MDR. However, genes encoding MBL such as IMP, VIM, NDM and SIM also have to be considered. With that in mind, a real-time PCR with TaqMan probes capable of detecting the class D carbapenemases present in A. baumannii has been defined and validated for that very purpose. The diagnostic industry is working to try to implement this technology from a direct clinical specimen, rather than from the isolated colony, which would translate into a significant reduction in response times.
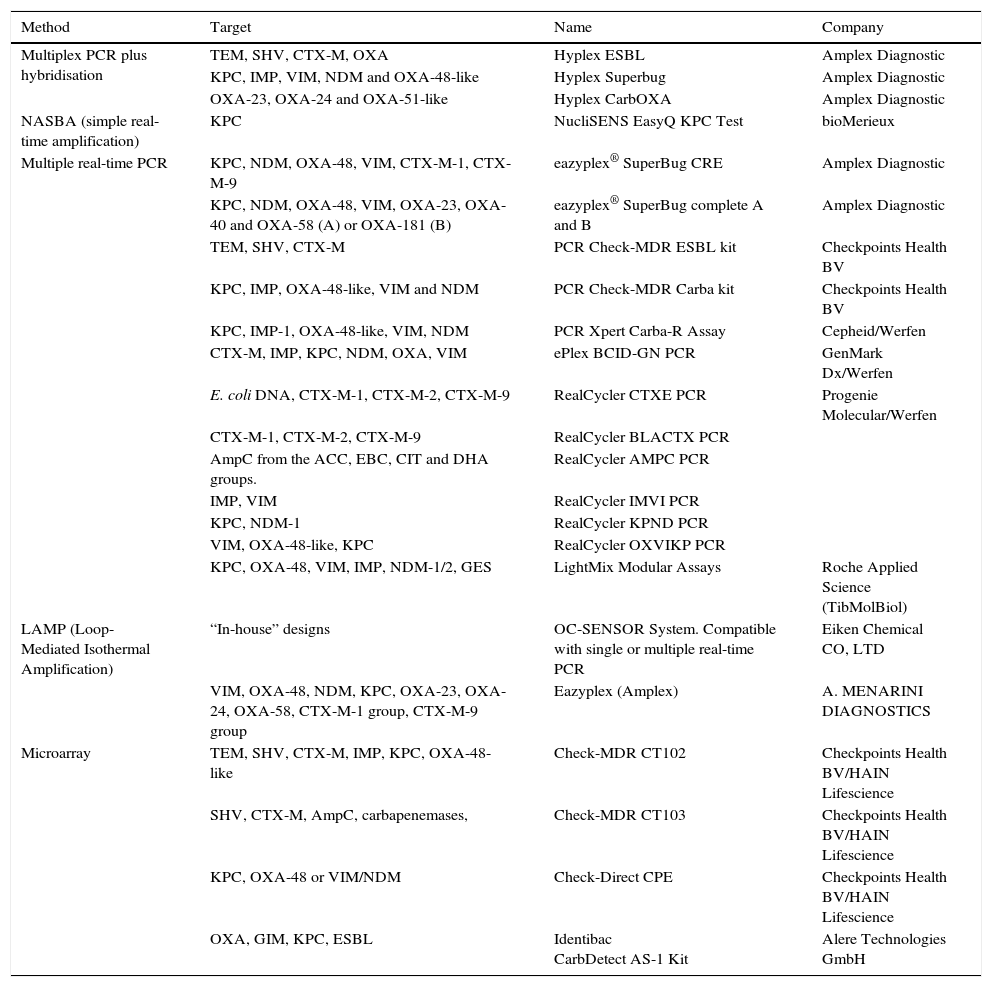
As mentioned above, numerous molecular methods have been marketed in recent years for the rapid detection of ESBL, AmpC and carbapenemase in rectal samples; a summary of some of the main commercial methods available is provided in Table 3.
Commercial methods for the molecular detection of ESBL, pAmpC and carbapenemase in rectal samples.
| Method | Target | Name | Company |
|---|---|---|---|
| Multiplex PCR plus hybridisation | TEM, SHV, CTX-M, OXA | Hyplex ESBL | Amplex Diagnostic |
| KPC, IMP, VIM, NDM and OXA-48-like | Hyplex Superbug | Amplex Diagnostic | |
| OXA-23, OXA-24 and OXA-51-like | Hyplex CarbOXA | Amplex Diagnostic | |
| NASBA (simple real-time amplification) | KPC | NucliSENS EasyQ KPC Test | bioMerieux |
| Multiple real-time PCR | KPC, NDM, OXA-48, VIM, CTX-M-1, CTX-M-9 | eazyplex® SuperBug CRE | Amplex Diagnostic |
| KPC, NDM, OXA-48, VIM, OXA-23, OXA-40 and OXA-58 (A) or OXA-181 (B) | eazyplex® SuperBug complete A and B | Amplex Diagnostic | |
| TEM, SHV, CTX-M | PCR Check-MDR ESBL kit | Checkpoints Health BV | |
| KPC, IMP, OXA-48-like, VIM and NDM | PCR Check-MDR Carba kit | Checkpoints Health BV | |
| KPC, IMP-1, OXA-48-like, VIM, NDM | PCR Xpert Carba-R Assay | Cepheid/Werfen | |
| CTX-M, IMP, KPC, NDM, OXA, VIM | ePlex BCID-GN PCR | GenMark Dx/Werfen | |
| E. coli DNA, CTX-M-1, CTX-M-2, CTX-M-9 | RealCycler CTXE PCR | Progenie Molecular/Werfen | |
| CTX-M-1, CTX-M-2, CTX-M-9 | RealCycler BLACTX PCR | ||
| AmpC from the ACC, EBC, CIT and DHA groups. | RealCycler AMPC PCR | ||
| IMP, VIM | RealCycler IMVI PCR | ||
| KPC, NDM-1 | RealCycler KPND PCR | ||
| VIM, OXA-48-like, KPC | RealCycler OXVIKP PCR | ||
| KPC, OXA-48, VIM, IMP, NDM-1/2, GES | LightMix Modular Assays | Roche Applied Science (TibMolBiol) | |
| LAMP (Loop-Mediated Isothermal Amplification) | “In-house” designs | OC-SENSOR System. Compatible with single or multiple real-time PCR | Eiken Chemical CO, LTD |
| VIM, OXA-48, NDM, KPC, OXA-23, OXA-24, OXA-58, CTX-M-1 group, CTX-M-9 group | Eazyplex (Amplex) | A. MENARINI DIAGNOSTICS | |
| Microarray | TEM, SHV, CTX-M, IMP, KPC, OXA-48-like | Check-MDR CT102 | Checkpoints Health BV/HAIN Lifescience |
| SHV, CTX-M, AmpC, carbapenemases, | Check-MDR CT103 | Checkpoints Health BV/HAIN Lifescience | |
| KPC, OXA-48 or VIM/NDM | Check-Direct CPE | Checkpoints Health BV/HAIN Lifescience | |
| OXA, GIM, KPC, ESBL | Identibac CarbDetect AS-1 Kit | Alere Technologies GmbH |
Several of these techniques have proven effective on clinical specimens other than rectal swabs.
The detection by molecular methods of a gene encoding for a broad-spectrum β-lactamase on a clinical specimen should be reported specifying the method used: “Presence of a gene encoding [resistance mechanism] detected using [method used]”. The species is not mentioned as it is not identified with this approach. In the case of a previously identified bacterial colony, the species should also be reported, “[Resistance mechanism]-producing [name of species] isolated”. Depending on the mechanism and the technique used, it is possible to specify the specific β-lactamase detected or the group or family to which it belongs. Detection of β-lactamase belonging to a group of ESBL, pAmpC or carbapenemases will be reported as, “Presence of a gene encoding [resistance-mechanism group/family] detected by [method used]”. Lack of detection will be reported as, “No presence of genes encoding [resistance mechanisms tested] detected using [method used]”.
Conflicts of interestThe authors declare that they have no conflicts of interest.
Please cite this article as: Oteo J, Bou G, Chaves F, Oliver A. Métodos microbiológicos para la vigilancia del estado de portador de bacterias multirresistentes. Enferm Infecc Microbiol Clin. 2017;35:667–675.