Serratia marcescens (SM) may cause nosocomial outbreaks in Neonatal Intensive Care Units (NICU). We describe an outbreak of SM in a NICU and propose additional prevention and control recommendations.
MethodsBetween March 2019 and January 2020, samples were taken from patients in the NICU (rectal, pharyngeal, axillary and other locations) and from 15 taps and their sinks. Control measures were implemented including thorough cleaning of incubators, health education to staff and neonates’relatives, and use of single-dose containers. PFGE was performed in 19 isolates from patients and in 5 environmental samples.
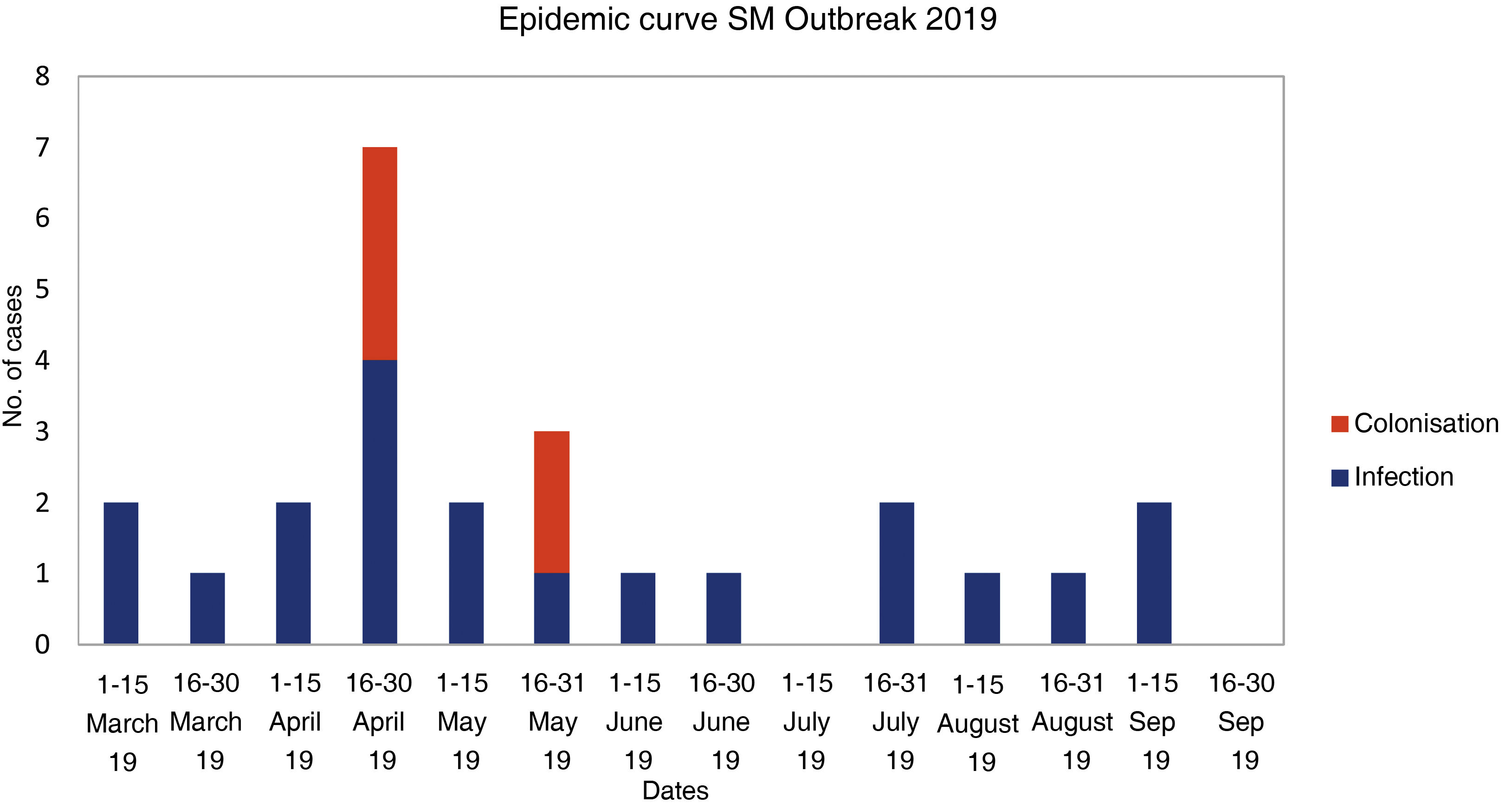
ResultsFrom the first case in March 2019 to the detection of the outbreak, a month elapsed. Finally, 20 patients were infected and 5 colonized. 80% of infected neonates had conjunctivitis, 25% bacteremia, 15% pneumonia, 5% wound infection, and 5% urinary tract infection. Six neonates had two foci of infection. Among the 19 isolates studied, 18 presented the same pulsotype and only one of the isolates from the sinkhole showed a clonal relationship with those of the outbreak. Initial measures established were ineffective to control de outbreak and were implemented with exhaustive cleaning, use of individual eye drops, environmental sampling and changing sinks.
ConclusionThis outbreak presented a high number of neonates affected due to its late detection and torpid evolution. The microorganisms isolated from the neonates were related to an environmental isolate. Additional prevention and control measures are proposed, including routine weekly microbiological sampling.
Serratia marcescens (SM) puede producir brotes nosocomiales en las Unidades de Cuidados Intensivos Neonatales (UCIN). Describimos un brote por SM en una UCIN y proponemos recomendaciones adicionales de prevención y control.
MétodosEntre marzo de 2019 y enero de 2020 se tomaron muestras en pacientes de la UCIN (frotis rectales, faríngeos, axilares y otras localizaciones) y de 15 grifos y sus sumideros. Se implementaron medidas de control incluyendo la limpieza exhaustiva de incubadoras, educación sanitaria a personal y familiares de neonatos, y uso de envases unidosis. Se hizo PFGE en 19 aislados de pacientes y en 5 muestras ambientales.
ResultadosDesde el primer caso (marzo 2019) hasta la detección del brote transcurrió un mes. Finalmente, 20 pacientes estaban infectados y 5 colonizados. El 80% de los neonatos infectados presentó conjuntivitis, 25% bacteriemia, 15% neumonía, 5% infección de herida y 5% infección urinaria. Seis neonatos tuvieron dos focos de infección. De los 19 aislados estudiados, 18 pertenecieron al mismo pulsotipo y sólo uno de los aislados del sumidero presentó relación clonal con los del brote. Las medidas iniciales para el control del brote fueron ineficaces y se implementaron con la limpieza exhaustiva de la unidad, uso de colirios individuales, toma de muestras ambientales y cambios de lavabos.
ConclusiónEste brote presentó un elevado número de neonatos afectados por su tardía detección y evolución tórpida. Las cepas de los pacientes estaban relacionadas con un aislado ambiental. Se proponen medidas adicionales de prevención y control de brotes incluyendo la toma de muestra microbiológica rutinaria semanal.
The hospital environment plays a major role in transmitting pathogens through cross-transmission,1 increasing healthcare-associated infections. Serratia marcescens (SM) is known for its ubiquity and can develop resistance to third-generation cephalosporins.2 In recent years it has been associated with outbreaks in neonatal intensive care units (NICUs).1 Some risk factors identified for colonisation/infection by this microorganism in the NICU are low birth weight, prematurity, long hospital stay, and the use of antibiotics and invasive measures.3
An SM outbreak in an NICU is a significant event, as it can spread rapidly.4 Its early detection is beneficial to prevent the spread.5,6 There is no universal consensus on the specific measures to implement in outbreaks of SM, and most studies reiterate similar actions; reinforcing hand hygiene, transmission-based contact precautions, health education of professionals and cohort isolation of infected or colonised patients.4 However, other factors, such as contamination of medical supplies, may cause the outbreak to spread.7,8 Microbiological tools such as pulsed, field electrophoresis (PFGE) and massive genomic sequencing make it possible to establish clonal relationships, and these apply to the study of outbreaks regarding their control.9,10 Nevertheless, these proposed measures may be insufficient in clinical practice for multiple reasons, such as environmental reservoirs.
This work aims to describe the evolution of an outbreak due to SM, the measures for its control and eradication, and the microbiological techniques used to review and propose new evaluation and control measures.
Material and methodsSiteThis was a cross-sectional, descriptive and retrospective study of an outbreak in the NICU of the Hospital Universitario Virgen de las Nieves (HUVN) [Virgen de las Nieves University Hospital], Granada, between March 2019 and January 2020. This NICU has 38 beds, of which 20 belong to the basic care module, 10 to the intermediate care module and 8 to the intensive care module (Supplementary Fig. S1 ). Its nursing staff then consisted of 39 nurses and 30 auxiliary nursing technicians.
Detection of the outbreakIn April 2019, the Microbiology Service reported the isolation of SM in a sample of conjunctival exudate from a neonate in the NICU to Preventive Medicine, with a warning that there seemed to be an aggregation of cases in recent months. An investigation began, with isolates requested from the NICU from January to April 2019, with seven cases (five conjunctival exudates and two blood cultures). On 26 April 2019, the Preventive Medicine Service declared an outbreak of SM in the NICU, which was reported to the Andalusian Epidemiological Surveillance System. Finally, 25 cases were detected, with the first isolation identified in a conjunctival exudate from 13 March 2019 (case 1) and the last from a conjunctival exudate from 12 September 2019 (case 25). The outbreak was concluded in January 2020, two months after case 20 was discharged.
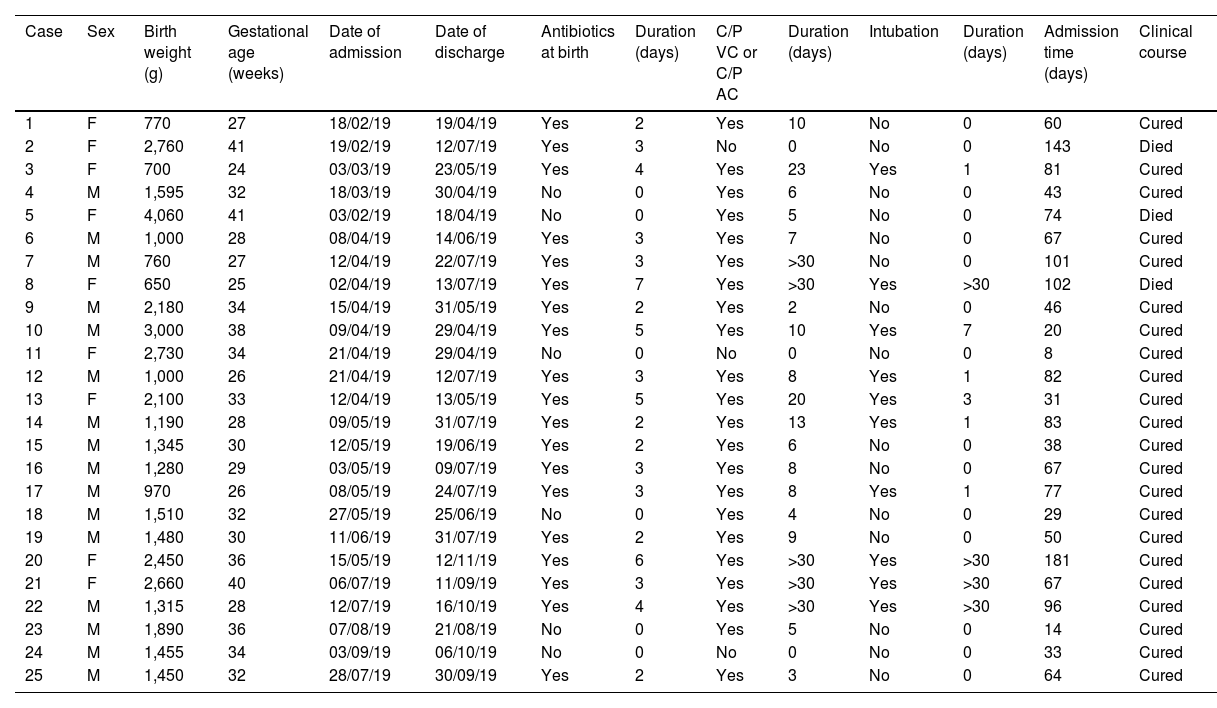
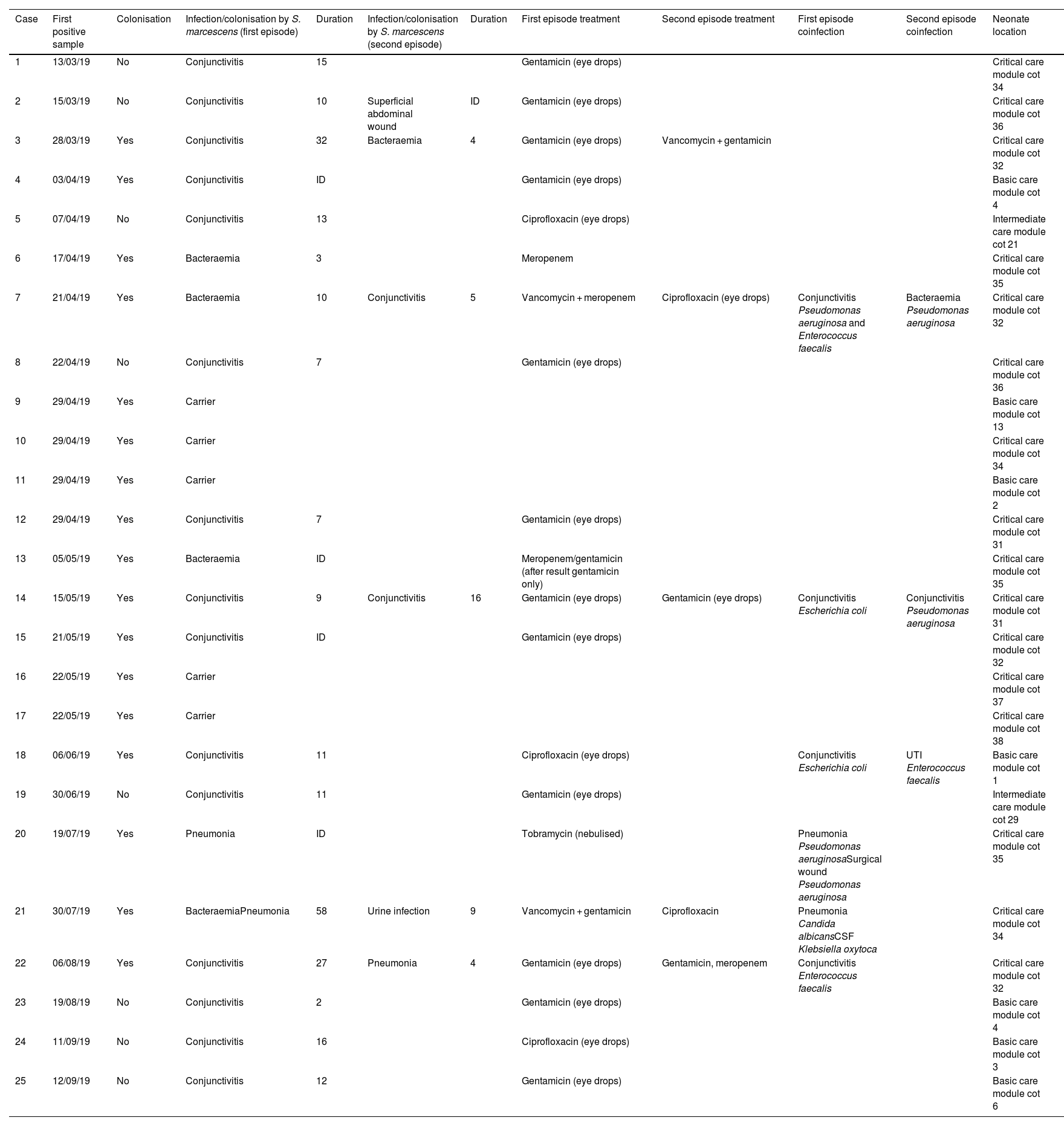
Clinical and epidemiological investigationThe microbiological screening was performed on neonates admitted to the NICU (29/04/19, 13/05/19 and 22/05/19) and in environmental reservoirs (09/08/19; 15 taps and 15 sink drains).11 A "case" was considered to be any neonate with isolation of SM admitted to the NICU during the outbreak period, whether from a clinical sample or from a study of carriers (rectal, pharyngeal, or axillary swabs). Those patients who had a sample which was positive for the study of carriers and who did not present with signs or symptoms of infection were categorised as "colonised"; and "infected" patients were those who had at least one clinical sample of any origin in which SM was isolated together with compatible symptoms. General clinical-demographic data (Table 1) and those related to the presence of SM (Table 2) were collected.
Intrinsic and extrinsic risk factors in neonates with infection/colonisation bySerratia marcescens admitted to the NICU.
| Case | Sex | Birth weight (g) | Gestational age (weeks) | Date of admission | Date of discharge | Antibiotics at birth | Duration (days) | C/P VC or C/P AC | Duration (days) | Intubation | Duration (days) | Admission time (days) | Clinical course |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 770 | 27 | 18/02/19 | 19/04/19 | Yes | 2 | Yes | 10 | No | 0 | 60 | Cured |
| 2 | F | 2,760 | 41 | 19/02/19 | 12/07/19 | Yes | 3 | No | 0 | No | 0 | 143 | Died |
| 3 | F | 700 | 24 | 03/03/19 | 23/05/19 | Yes | 4 | Yes | 23 | Yes | 1 | 81 | Cured |
| 4 | M | 1,595 | 32 | 18/03/19 | 30/04/19 | No | 0 | Yes | 6 | No | 0 | 43 | Cured |
| 5 | F | 4,060 | 41 | 03/02/19 | 18/04/19 | No | 0 | Yes | 5 | No | 0 | 74 | Died |
| 6 | M | 1,000 | 28 | 08/04/19 | 14/06/19 | Yes | 3 | Yes | 7 | No | 0 | 67 | Cured |
| 7 | M | 760 | 27 | 12/04/19 | 22/07/19 | Yes | 3 | Yes | >30 | No | 0 | 101 | Cured |
| 8 | F | 650 | 25 | 02/04/19 | 13/07/19 | Yes | 7 | Yes | >30 | Yes | >30 | 102 | Died |
| 9 | M | 2,180 | 34 | 15/04/19 | 31/05/19 | Yes | 2 | Yes | 2 | No | 0 | 46 | Cured |
| 10 | M | 3,000 | 38 | 09/04/19 | 29/04/19 | Yes | 5 | Yes | 10 | Yes | 7 | 20 | Cured |
| 11 | F | 2,730 | 34 | 21/04/19 | 29/04/19 | No | 0 | No | 0 | No | 0 | 8 | Cured |
| 12 | M | 1,000 | 26 | 21/04/19 | 12/07/19 | Yes | 3 | Yes | 8 | Yes | 1 | 82 | Cured |
| 13 | F | 2,100 | 33 | 12/04/19 | 13/05/19 | Yes | 5 | Yes | 20 | Yes | 3 | 31 | Cured |
| 14 | M | 1,190 | 28 | 09/05/19 | 31/07/19 | Yes | 2 | Yes | 13 | Yes | 1 | 83 | Cured |
| 15 | M | 1,345 | 30 | 12/05/19 | 19/06/19 | Yes | 2 | Yes | 6 | No | 0 | 38 | Cured |
| 16 | M | 1,280 | 29 | 03/05/19 | 09/07/19 | Yes | 3 | Yes | 8 | No | 0 | 67 | Cured |
| 17 | M | 970 | 26 | 08/05/19 | 24/07/19 | Yes | 3 | Yes | 8 | Yes | 1 | 77 | Cured |
| 18 | M | 1,510 | 32 | 27/05/19 | 25/06/19 | No | 0 | Yes | 4 | No | 0 | 29 | Cured |
| 19 | M | 1,480 | 30 | 11/06/19 | 31/07/19 | Yes | 2 | Yes | 9 | No | 0 | 50 | Cured |
| 20 | F | 2,450 | 36 | 15/05/19 | 12/11/19 | Yes | 6 | Yes | >30 | Yes | >30 | 181 | Cured |
| 21 | F | 2,660 | 40 | 06/07/19 | 11/09/19 | Yes | 3 | Yes | >30 | Yes | >30 | 67 | Cured |
| 22 | M | 1,315 | 28 | 12/07/19 | 16/10/19 | Yes | 4 | Yes | >30 | Yes | >30 | 96 | Cured |
| 23 | M | 1,890 | 36 | 07/08/19 | 21/08/19 | No | 0 | Yes | 5 | No | 0 | 14 | Cured |
| 24 | M | 1,455 | 34 | 03/09/19 | 06/10/19 | No | 0 | No | 0 | No | 0 | 33 | Cured |
| 25 | M | 1,450 | 32 | 28/07/19 | 30/09/19 | Yes | 2 | Yes | 3 | No | 0 | 64 | Cured |
C/P AC, central or peripheral arterial catheter; C/P VC, central or peripheral venous catheter; F, female; M, male; NICU, Neonatal Intensive Care Unit.
Timing, types of infection/colonisation, treatments and location of neonates with Serratia marcescens admitted to the NICU.
| Case | First positive sample | Colonisation | Infection/colonisation by S. marcescens (first episode) | Duration | Infection/colonisation by S. marcescens (second episode) | Duration | First episode treatment | Second episode treatment | First episode coinfection | Second episode coinfection | Neonate location |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 13/03/19 | No | Conjunctivitis | 15 | Gentamicin (eye drops) | Critical care module cot 34 | |||||
| 2 | 15/03/19 | No | Conjunctivitis | 10 | Superficial abdominal wound | ID | Gentamicin (eye drops) | Critical care module cot 36 | |||
| 3 | 28/03/19 | Yes | Conjunctivitis | 32 | Bacteraemia | 4 | Gentamicin (eye drops) | Vancomycin + gentamicin | Critical care module cot 32 | ||
| 4 | 03/04/19 | Yes | Conjunctivitis | ID | Gentamicin (eye drops) | Basic care module cot 4 | |||||
| 5 | 07/04/19 | No | Conjunctivitis | 13 | Ciprofloxacin (eye drops) | Intermediate care module cot 21 | |||||
| 6 | 17/04/19 | Yes | Bacteraemia | 3 | Meropenem | Critical care module cot 35 | |||||
| 7 | 21/04/19 | Yes | Bacteraemia | 10 | Conjunctivitis | 5 | Vancomycin + meropenem | Ciprofloxacin (eye drops) | Conjunctivitis Pseudomonas aeruginosa and Enterococcus faecalis | Bacteraemia Pseudomonas aeruginosa | Critical care module cot 32 |
| 8 | 22/04/19 | No | Conjunctivitis | 7 | Gentamicin (eye drops) | Critical care module cot 36 | |||||
| 9 | 29/04/19 | Yes | Carrier | Basic care module cot 13 | |||||||
| 10 | 29/04/19 | Yes | Carrier | Critical care module cot 34 | |||||||
| 11 | 29/04/19 | Yes | Carrier | Basic care module cot 2 | |||||||
| 12 | 29/04/19 | Yes | Conjunctivitis | 7 | Gentamicin (eye drops) | Critical care module cot 31 | |||||
| 13 | 05/05/19 | Yes | Bacteraemia | ID | Meropenem/gentamicin (after result gentamicin only) | Critical care module cot 35 | |||||
| 14 | 15/05/19 | Yes | Conjunctivitis | 9 | Conjunctivitis | 16 | Gentamicin (eye drops) | Gentamicin (eye drops) | Conjunctivitis Escherichia coli | Conjunctivitis Pseudomonas aeruginosa | Critical care module cot 31 |
| 15 | 21/05/19 | Yes | Conjunctivitis | ID | Gentamicin (eye drops) | Critical care module cot 32 | |||||
| 16 | 22/05/19 | Yes | Carrier | Critical care module cot 37 | |||||||
| 17 | 22/05/19 | Yes | Carrier | Critical care module cot 38 | |||||||
| 18 | 06/06/19 | Yes | Conjunctivitis | 11 | Ciprofloxacin (eye drops) | Conjunctivitis Escherichia coli | UTI Enterococcus faecalis | Basic care module cot 1 | |||
| 19 | 30/06/19 | No | Conjunctivitis | 11 | Gentamicin (eye drops) | Intermediate care module cot 29 | |||||
| 20 | 19/07/19 | Yes | Pneumonia | ID | Tobramycin (nebulised) | Pneumonia Pseudomonas aeruginosaSurgical wound Pseudomonas aeruginosa | Critical care module cot 35 | ||||
| 21 | 30/07/19 | Yes | BacteraemiaPneumonia | 58 | Urine infection | 9 | Vancomycin + gentamicin | Ciprofloxacin | Pneumonia Candida albicansCSF Klebsiella oxytoca | Critical care module cot 34 | |
| 22 | 06/08/19 | Yes | Conjunctivitis | 27 | Pneumonia | 4 | Gentamicin (eye drops) | Gentamicin, meropenem | Conjunctivitis Enterococcus faecalis | Critical care module cot 32 | |
| 23 | 19/08/19 | No | Conjunctivitis | 2 | Gentamicin (eye drops) | Basic care module cot 4 | |||||
| 24 | 11/09/19 | No | Conjunctivitis | 16 | Ciprofloxacin (eye drops) | Basic care module cot 3 | |||||
| 25 | 12/09/19 | No | Conjunctivitis | 12 | Gentamicin (eye drops) | Basic care module cot 6 |
CSF: cerebrospinal fluid; NICU: Neonatal Intensive Care Unit; UTI: urinary tract infection.
These measures were the support of the hospital management, staff education with periodic and onboarding training, appropriate use of antibiotic therapy, epidemiological surveillance and the use of and adherence to transmission-based precautions, environmental measures and, finally, decolonisation, only in the event of a nosocomial outbreak if deemed necessary. Due to the ubiquity and the form of transmission of SM, thorough cleaning and disinfection of incubators and cohort isolation/grouping in patients colonised/infected by MS are implemented, with exclusive staff for them, and, ultimately, closure of the unit if the outbreak is not controlled, a measure that did not have to be undertaken in the outbreak described. Measures were carried out to control the outbreak, detailed by month of implementation (Appendix B Supplementary material).4
Microbiological study and clonal relationship analysisThe isolates were identified using MALDI-TOF (Biotyper, Bruker Daltonics, Billerica, MA, USA), and the antimicrobial susceptibility study was conducted on clinical isolates using MicroScan WalkAway (Beckman-Coulter, Brea, CA, USA.). For the study of the epidemiological relationship at the molecular level, a single isolate per patient and all the isolates detected in environmental samples were selected the isolates detected in environmental samples. They were sent to the reference laboratory of the Programa Andaluz de Vigilancia y Control de Infecciones Asociadas a la Asistencia Sanitaria y Administración de Antibióticos [Andalusian Programme for Surveillance and Control of Healthcare-Associated Infections and Administration of Antibiotics] (PIRASOA programme). Antimicrobial susceptibility tests were performed in this laboratory by microdilution and were interpreted by EUCAST (https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Consultation/2018/Information_for_NACs_-_Redefining_antimicrobial_susceptibility_testing_categories.pdf). All isolates were analysed by PFGE with XbaI-DNA. (http://www.cdc.gov/pulsenet). Isolates that differed by two or more bands in PFGE assays were assigned to different pulsotypes. Isolates with no band differences (100% similarity) were considered identical; and isolates with more than five band differences (<80% similarity) were considered different. The resulting restriction fragments were separated on the CHEF DR-II system (Bio-Rad Laboratories, Alcobendas, Madrid, Spain) with 1% agarose gel. Subsequently, they were stained with ethidium bromide, illuminated with ultraviolet light, and photographed on a Gel Logic 200 Automated Imaging System (Kodak, Rochester, NY, USA). Band pattern conversion, normalisation and analysis was performed using BioNumerics 12.0 software (AppliedMaths, Jollyville Rd., Austin, TX, USA), analysing the patterns as described above, using the Dice coefficient with position tolerance settings of 1% optimisation and 1% band position tolerance. There is no MLST scheme developed by the Pasteur Institute to characterise this species of bacteria. However, all the isolates that presented the same pulsotype were related.
ORION declarationThe ORION statement guidelines were followed to ensure transparency in outbreak reporting (https://www.ucl.ac.uk/drupal/site_antimicrobial-resistance/sites/antimicrobial-resistance/files/checklist_authors.pdf).
Ethical approvalThe Granada Research Ethics Committee approved the project on 21/12/2021 with reference code 2390-N-21.
ResultsClinical-epidemiological researchOn 29 April, samples were taken from 19 neonate carriers (rectal and pharyngeal swabs); six were already classified as cases, and four new colonised neonates were identified. On 13 May, control samples were taken from five cases. Finally, on 22 May, samples were taken from eight neonates admitted, three of them previously being part of the outbreak, and two new colonised neonates identified. The status of one of the colonised patients changed to infection (conjunctivitis), and the outbreak was finally made up of 20 infected (80%) and five colonised patients (20%). The evolution of the cases can be observed in the epidemic curve (Fig. 1). Twenty-five patients were analysed, 16 (64%) male. Their weight and mean gestational age at birth were 1,692 g (range: 650−4,060 g and SD: 857.46) and 31.64 weeks (range: 24–41 weeks and SD: 5.02), respectively. The mean length of stay in the NICU was 66.28 days (range: 8–181 days and SD: 39.48) (Table 2). Twelve (60%) of the 20 neonates with infection were colonised at the rectal level. Of the 20 infected patients, 16 (80%) presented with conjunctivitis, five (25%) bacteraemia, three (15%) pneumonia, one (5%) surgical wound infection, and one (5%) urinary infection. SM was isolated in more than one location in six neonates (30% of those infected and 24% of the total), and one of them presented with two episodes of conjunctivitis. Thirteen cases of conjunctivitis were treated with gentamicin eye drops (76.5%) and four with ciprofloxacin (23.5%). In six neonates (30% of the 20), infection by another microorganism was simultaneously detected (Table 2). Three neonates (12%) died due to causes unrelated to SM infection. At the time of the first microbiological isolation (Table 2), 16 (64%) cases were in the critical care module, seven (28%) were in basic care, and two (8%) were in intermediate care. Regarding the environmental samples, 100% of the tap samples were negative. SM was detected in four sink drains (five isolates, 26.7%) (Table 3), located in the incubator laundry room (1, 25%), critical care (2, 50%) and intermediate care (1, 25%).
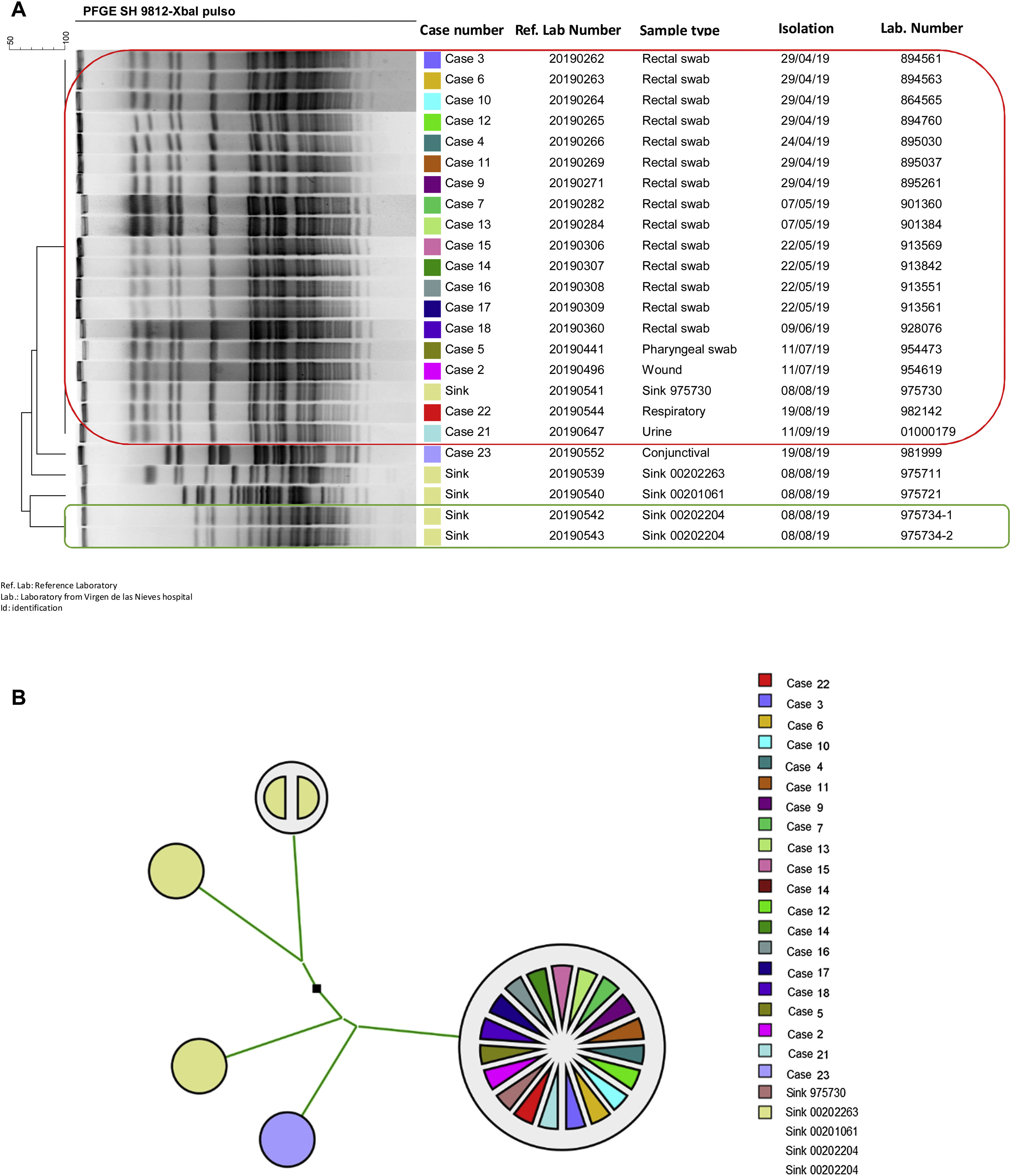
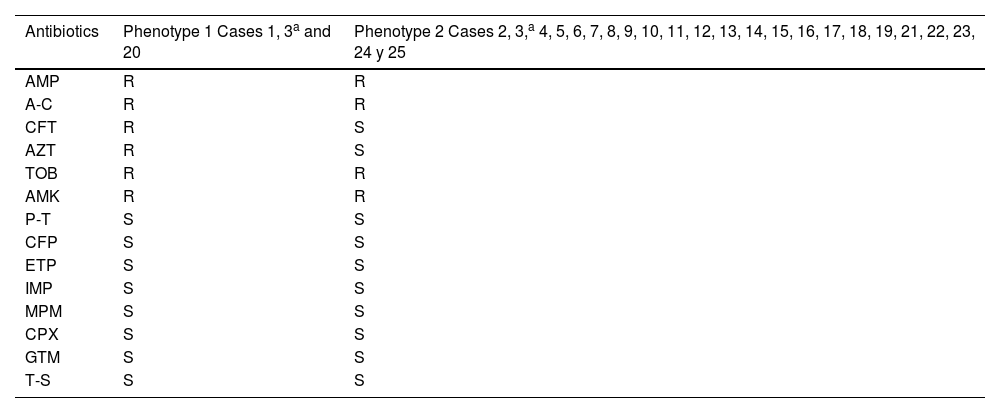
Two different susceptibility profiles were detected (Table 4). Of case three, only one isolate with phenotype two was analysed. Of the 25 cases defined in the outbreak, the clonal relationship of 19 clinical isolates was analysed (14 rectal swabs and one pharyngeal swab; one urine; one wound exudate, one respiratory and another conjunctival) and five from four environmental reservoirs. Ninety-four point seven per cent of the isolates detected in patient samples were identical; only one isolate (case 23) had a different pulsotype. One environmental isolate (sink drain 975730) was identical to this cluster of clinical isolates. The rest of the environmental isolates had different pulsotypes than the isolates from the outbreak. The two isolates detected in the same sink drain were identical to each other (Fig. 2).
Susceptibility profiles of Serratia marcescensisolates.
| Antibiotics | Phenotype 1 Cases 1, 3a and 20 | Phenotype 2 Cases 2, 3,a 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 21, 22, 23, 24 y 25 |
|---|---|---|
| AMP | R | R |
| A-C | R | R |
| CFT | R | S |
| AZT | R | S |
| TOB | R | R |
| AMK | R | R |
| P-T | S | S |
| CFP | S | S |
| ETP | S | S |
| IMP | S | S |
| MPM | S | S |
| CPX | S | S |
| GTM | S | S |
| T-S | S | S |
A-C, amoxicillin-clavulanic acid; AMK, amikacin; AMP, ampicillin; AZT, aztreonam; CFP, cefepime; CFT, cefotaxime; CPX, ciprofloxacin; ETP, ertapenem; GTM, gentamicin; IMP, imipenem; MPM, meropenem; P-T, piperacillin-tazobactam; R, resistant; S, susceptible; T-S, trimethoprim-sulfamethoxazole; TOB, tobramycin.
Clonal relationship of isolates ofSerratia marcescens from patient and environmental samples from the HUVN Neonatology Unit (Granada). (A) Dendrogram of isolates of S. marcescens sent to the PIRASOA reference laboratory. Identical isolates are shown in a box (red: 18 isolates from neonates and isolate from sink drain; green: isolates from the same sink drain). (B) Phylogenetic tree of isolates of S. marcescens sent to the PIRASOA reference laboratory.
Due to the prolonged duration of the SM nosocomial outbreak in the NICU, new measures were added to the initial actions for its control since cases continued to appear (Supplementary Table S1). After the implementation of all the outbreak control measures, to which was added control of antibiotic therapy, discharge of newborns when clinical conditions allowed for it, thorough clinical and environmental controls, and the involvement of all the NICU professionals, the outbreak was concluded two months after the last case, due to the possibility of SM persisting on inanimate surfaces for several months.
DiscussionSM has been identified as the third leading cause of outbreaks in hospitals,12 as in the NICU in children with low birth weight, prematurity, a long stay in the ICU and the presence of devices.13 In our study, most neonates had low birth weight, were premature, and had a catheter. In this outbreak, there were three (12%) deaths, none due to bacteraemia, which has been described previously.3,14 The high number of episodes compared to other studies stands out.15 This could be due to non-compliance with standard control measures and the late detection of the outbreak, which led to a delay in the initiation of control measures, since it wasn't until there were seven confirmed cases that it was notified, at the end of April 2019. Some studies have identified sources and reservoirs of SM during the outbreak investigation, for example, in disinfectants,16 parenteral nutrition,17 shampoos18 or water containers,19 but in most the source is not clear.20 In this study, case 1 was the first identified case with SM infection in March 2019, but we cannot be sure that it was the initial source, since there could have been neonates previously admitted who did not have carrier samples taken. In addition, it was impossible to study the pulsotype in this case. The presence of the microorganism was not analysed in the relatives of the neonates, which should also be taken into account. Although one of the measures implemented was health education (emphasising hand hygiene), no observations were performed. People entering and leaving the NICU could have introduced a microorganism into this ward. Hospital staff were not tested for SM either, but hand hygiene measures, transmission-based precautions, and proper discipline regarding clothing in a NICU were reinforced. This decision was made because conducting SM screening in the staff would not have contributed any relevant information. In fact, conducting a study of carriers in this sector is not recommended since they are rarely the cause.15,21 One of the control measures for the outbreak was reinforcing hand hygiene among hospital staff, especially NICU staff. To evaluate this, hand hygiene observations were performed between 15 May and 5 June, and the analysis revealed 62.4% compliance, somewhat higher than the average resulting from a systematic review (59.6%), although lower than that observed in other NICUs (67%).22
Regarding the transmission of the outbreak, as in others described, the presence of asymptomatic neonates colonised by SM through the hands of hospital staff 24,25 would constitute the main reservoir.23 In addition, as part of the outbreak control measures, an environmental sample was taken, and four contaminated sink drains were identified; the sink for incubators and other equipment for use in neonates was the only one identical to the isolates from the outbreak. This fact does not imply that this is the source of the outbreak since the fact that it was a sink drain means it could have been contaminated after washing the incubators where the neonates resided ("environmental case"). However, it is a factor in the spread of the outbreak since SM can remain on inanimate surfaces for months.26 After changing the taps and sinks, cases continued to appear, which supports the fact that the neonates themselves were the main reservoir, and the staff, transmission vectors, and sink drains continued to be colonised after using the sinks whilst positive cases were present.
In addition, the appearance of new cases may be related to the lower nursing/patient ratio and a deficiency in the training of professionals in prevention measures20 since between June and September, there were fewer staff and/or there were fewer specialised staff working in an open room, which does not facilitate hand hygiene. Other relevant factors regarding the spread of the outbreak were the age of the NICU and its layout: reduced space between incubators and flowing traffic of professionals from different specialties, relatives of neonates, and transfer of patients between the three modules.
Regarding the samples taken from neonates and sent to the reference laboratory, there was one case (case 23) in which, after analysis, it was observed that SM belonged to a different pulsotype from that of the outbreak. It is noteworthy that in this patient, the sample was taken 13 days after the last case detected (case date identified hitherto: 06/08/19), and it was also found in a different location than the last cases, which points to the introduction of a new SM lineage in the NICU. The isolates from cases 24 and 25 were not analysed using PFGE, so it is impossible to confirm whether SM belonged to the same epidemiological chain of the outbreak or another had arisen from case 23. Also, identifying other lineages in other sink drains supports the hypothesis that there are different strains, with different chains of transmission, that coincide in time during this nosocomial outbreak.
Despite all the control measures undertaken in this outbreak, NICU outbreaks are not uncommon to continue, just as they did in November 2020. In fact, other reports have identified secondary and tertiary outbreaks.27–29 Based on the above, the following proposals for improvement are made to be implemented to avoid the consequences that a similar outbreak can cause in a NICU. First, constant surveillance in high-risk units and effective communication between hospital services in the event of suspected nosocomial infection events are important to detect outbreaks early and thus act promptly to prevent spread in the hospital environment. Secondly is microbiological screening as part of the clinical routine, with a systematic periodicity (at least biweekly), for detecting "unusual" microorganisms early. In this way, the transmission of the microorganism can be interrupted early, and outbreaks with a high number of cases can be avoided. According to a German study, screening all neonates admitted to the NICU is useful for the early detection of these microorganisms.30 Further, we believe that a consensus document on universal measures for action in outbreaks would be valuable, in addition to the need for continuous training of all hospital professionals, especially those who attend high-risk units, in measures to prevent nosocomial infection, emphasising hand hygiene.
Regarding the limitations of this study, we would emphasise that pulsotypes were not determined for all the outbreak cases; specifically, from case 3, several isolates with phenotypes 1 and 2 were obtained. However, the molecular analysis could only be performed on phenotype 2. Another aspect is the fact that, mainly due to the lack of human resources, screening was not continued, which could have contributed to the persistence of the outbreak for a longer period, as well as the fact that environmental reservoirs were not analysed until several months after the detection of the outbreak.
In conclusion, an outbreak of SM occurred in a NICU of a regional hospital in south-eastern Spain, where there was a high number of affected individuals due to its late detection and persistence. In addition to the usual prevention and control measures, new measures are proposed, such as scheduled, routine microbiological screening for prevention and control, as well as continuous training of staff in hygiene measures, emphasising hand hygiene.
FundingThe authors declare that they have not received any funding to carry out this work.
Conflicts of interestThe authors declare that they have no conflicts of interest