Resistance-associated substitutions (RASs) to the new HCV NS5A inhibitor elbasvir may limit its efficacy and lead to virological failure in HCV-GT1a-infected patients. There are no data outside clinical trials evaluating their prevalence and impact in grazoprevir/elbasvir in GT1a-infected patients in Spain. A multicentre cross-sectional study of 632 initial patients was conducted. In 13 of these patients, the sample could not be amplified or a consensus sequence by Sanger sequencing could not be performed. Ultimately, 617 HCV-G1a-infected individuals treated at 84 Spanish hospitals from the 17 autonomous communities plus the 2 autonomous cities of Spain were analysed. HCV population sequencing was used to identify RAS to elbasvir and the mutational pattern and drug sensitivity were confirmed by geno2pheno[HCV]. Viruses bearing RASs to elbasvir were present in 6.2% of HCV-G1a infected patients. The most common RASs were the Y93C/H/N and Q30E/H/R (2.4% and 2.3%, respectively). Only 3.4% of the identified RASs to elbasvir conferred reduced susceptibility to elbasvir by geno2pheno[HCV], which exclusively identified the positions Q30H/R (n=7) and Y93C/H/N (n=8) as single mutations and Q30H+Y93H (n=4) and Q30R+Y93H (n=2) as double mutations as the major RASs to elbasvir. A lower prevalence of RASs to elbasvir was observed in our HCV-G1a Spanish cohort than reported previously in clinical trials evaluating patients from the USA. This information may be essential to guide the implementation of grazoprevir/elbasvir in Spain and to manage G1a-infected patients.
Las sustituciones asociadas a resistencia (RAS) a elbasvir, el nuevo inhibidor de la NS5A del virus de la hepatitis C (VHC), pueden presentar relevancia al limitar su eficacia y conducir al fracaso virológico en pacientes infectados por VHC genotipo 1a (GT1a) a diferencia de lo observado en GT1b y GT4. No existen datos fuera de ensayos clínicos que evalúen la prevalencia y el impacto de grazoprevir/elbasvir en pacientes infectados con GT1a en España. Se llevó a cabo un estudio transversal multicéntrico en 632 pacientes iniciales, en 13 de los cuales no se consiguió amplificar la muestra o no fue válida para alcanzar una secuencia consenso mediante secuenciación de Sanger. Finalmente, se analizaron 617 individuos infectados con VHC-GT1a atendidos en 84 hospitales distribuidos por las 17 comunidades autónomas más las 2 ciudades autónomas que conforman el territorio español. La población de VHC secuenciada se ha usado para identificar RAS a elbasvir, mientras que el patrón mutacional y la sensibilidad farmacológica se confirmaron mediante geno2pheno[HCV]. Los virus portadores de RAS a elbasvir se observaron en el 6,2% de los pacientes infectados con el VHC-G1a. Las RAS más comunes fueron Y93C/H/N y Q30E/H/R (2,4 y 2,3%, respectivamente). Solo el 3,4% de las RAS a elbasvir identificadas confirieron susceptibilidad reducida al fármaco mediante geno2pheno[HCV] que identificó exclusivamente como principales RAS a elbasvir las posiciones Q30H/R (n=7) y Y93C/H/N (n=8) como mutaciones simples y Q30H+Y93H (n=4) y Q30R+Y93H (n=2) como mutaciones dobles. En nuestra cohorte española con VHCG1a se observó una menor prevalencia de RAS a elbasvir que la reportada previamente en ensayos clínicos realizados en pacientes norteamericanos. Esta información puede ser esencial para el manejo de los pacientes infectados con G1a y guiar la implementación de grazoprevir/elbasvir en España.
Grazoprevir/elbasvir (EBR) was approved by the Food and Drug Administration in January 2016.1 In the European Union, the European Medicines Agency approved it in July 2016, and it was included in the list of medicines under additional monitoring.2 In Spain, the Ministry of Health approved public funding for this drug on 1 September 2016. This drug is the latest combination product of direct-acting antivirals (DAAs) for the treatment of hepatitis C virus (HCV) to be incorporated into the therapeutic arsenal and has been approved for use in GT1 and GT4.1 The use of grazoprevir/EBR is not indicated in patients infected with genotypes 2, 3, 5 and 6. Grazoprevir belongs to the family of HCV NS3/4A protease inhibitors and EBR is the first second-generation HCV NS5A inhibitor. However, the presence of resistance-associated substitutions (RASs) may reduce sensitivity to DAAs, limiting their efficacy. Previous studies have demonstrated that the presence of RASs in the NS3 region does not affect the efficacy of grazoprevir in patients not previously treated with new DAAs or patients who have never received treatment.3 However, RASs in the NS5A region have a strong impact on the efficacy of EBR.
The impact of pre-existing HCV variants on the response to therapy with grazoprevir/EBR in subjects infected with HCV who have never received treatment was evaluated in the C-EDGE TN study, in which RASs to EBR were identified in 12% (19/154) of participants carrying genotype 1a (GT1a) and associated with a 58% (11/19) reduction in sustained viral response at 12 weeks.3 The recommendation to extend treatment with grazoprevir/EBR to 16 weeks with the addition of ribavirin for never-treated HCV-GT1a patients with NS5A RASs is based on the results of the C-EDGE TE study.4 Specifically, NS5A base variants with a 5 times greater change to EBR reduced the sustained viral response at 12 weeks to 52.4% (11/21) in subjects infected with HCV-GT1a, while sustained viral response at 12 weeks increased by 94.8% (55/58) in the intention-to-treat analysis when ribavirin was added to grazoprevir/EBR and treatment extended to 16 weeks. In patients infected with HCV-GT1b and HCV-GT4, the efficacy of grazoprevir/EBR was 99% and 100%, respectively3.
The failure of therapy with EBR in patients infected with HCV-GT1a has been associated with the presence of the specific substitutions M28A/G/T, Q30D/E/G/H/K/L/R. L31F/M/V. H58D and Y93C/H/N/S in accordance with the 2016 European Recommendations,5 which represent a subset of NS5A RASs.6 Cross-resistance has been demonstrated between NS5A RASs to EBR and approved first-generation NS5A inhibitors,6,7 as has the high persistence of these substitutions in the viral population once established.8,9 Both phenomena could limit the use of the NS5A inhibitors family. Based on this problem, the study of NS5A resistances in patients infected with HCV-GT1a who are candidates for treatment with grazoprevir/EBR is recommended10.
On 1 September 2016 in Spain, the Ministry of Health approved public funding for this drug and on 3 February 2017, the Spanish Agency of Medicines and Medical Devices published its Therapeutic Positioning Report on EBR and grazoprevir (Zepatier®) in chronic hepatitis C, thereby beginning its official establishment.11 The HCV-GT1a genotype represents approximately 23% of the genotypes circulating in Spain.12 Prior knowledge of the presence of NS5A RASs to EBR in the Spanish population infected with HCV-GT1a would therefore help to identify the proportion of patients who might benefit from this new treatment. This study provides an analysis of RASs through Sanger gene sequencing in Spanish patients infected with HCV-GT1a in order to evaluate the prevalence and impact of NS5A RASs of clinical relevance to the new NS5A inhibitor drug.
MethodsStudy Design and PatientsA multi-centre, cross-sectional study was conducted on 632 individuals with chronic HCV-GT1a infection who had never received treatment with NS5A inhibitors. The STROBE verification list was used to help design and direct the study13.
The patients were treated in 84 Spanish health centres distributed throughout the country (see Appendix A, additional data). The samples were selected randomly and were collected between October 2014 and February 2015, before starting anti-NS5A therapy. The plasma specimens were sent together with a minimum set of data to the Viral Infection and Immunity Unit of the Viral Hepatitis Research and Reference Laboratory of the National Centre for Microbiology [Centro Nacional de Microbiología] (Instituto de Salud Carlos III [ISCIII] - the main Public Biomedical Research Body in Spain) through the ISCIII's Diagnostic Orientation Area. Both data and samples were anonymised and transferred to the ISCIII's National Biobank (Ref.: B.0000984).
EthicsThe study was conducted in accordance with the Declaration of Helsinki. It was approved by the Institutional Review Board and the ISCIII's Independent Ethics Committee (CEI PI 43_2015).
NS5A Amplification and SequencingViral RNA was extracted from plasma using the QIAsymphony DSP Virus/Pathogen kit (Qiagen, Hilden, Germany) from 500 μl, and complete amplification of the NS5A gene was achieved with the RT-PCR OneStep kit (Qiagen, Hilden, Germany) using oligonucleotides NS4BFW (5′TGAGGCGACTVCACCAGTGG3′) and NS5BRV (5′TCTTCCGCGGCRCACGGGGTGA3′). The amplification protocol was: 30 min at 54°C; 15 min at 95°C; 35 repeated cycles of 30 sec at 94°C, 30 sec at 60°C and 2 min at 72°C in the Applied Biosystems VeritiTM thermal cycler. Positive and negative controls were included in all amplifications. Positive PCR products were visualised with GelRed (Biotium, USA) as a specific HCV band of ∼1.343 bp. The amplicons were purified (illustra™ GFX™ PCR DNA and Gel Band Purification Kit, GE Healthcare, USA) and diluted 1:2 in nuclease-free water (Roche). The sequencing reaction was then performed with the following oligonucleotides: FwSc2 (5′CGACTRCACCAGTGGATAAGC3′); FwSc3 (5′CTRCACCAGTGGATAAGCTCG3′); FwSc5 (5′CCCATTAACGCCTACACCACG3′); FwSc7 (5′CCTGACGCCGAGCTCATAGAG3′); RvSc3 (5′AGCGAGTGTGCATGATGCCAT3′); RvSc7 (5′GTGCGCCTGTCCAGGAATAAGA3′) and Sanger sequencing was performed using the ABI PRISM® 377 DNA sequencer (Applied Biosystems, Foster City, USA).
Bioinformatic AnalysisThe SeqMan (Lasergne DNASTAR Inc., Madison, WI, USA) program was used to obtain consensus sequences, while the NS5A sequences were aligned with the HCV-GT1a representative sequence H77 using the MEGA6 (Molecular Evolutionary Genetics Analysis Version 6.0; http://www.megasoftware.net/) program. The RAS detection cut-off point used was 15% in accordance with the European Recommendations, and the software used was Minor Variant Finder (Applied Biosystems). To determine the prevalence of NS5A RASs to EBR in patients with HCV-GT1a who had never received NS5A inhibitors, the NS5A gene was analysed in accordance with the latest European Recommendations.5 The NS5A mutation pattern and pharmacological sensitivity were confirmed using geno2pheno[HCV] (g2p[HCV]) (Bonn, Germany; http://hcv.geno2pheno.org/). HCV-GT1a lineages (clade i and clade ii) were identified using g2p[HCV]. Isolate H77 (GenBank access number AF009606) was used as a GT1a reference sequence. The level of resistance to EBR (copy number) of each RAS identified in our cohort was evaluated in accordance with Black et al. (2015)8 and Cento et al. (2015)14.
Statistical AnalysisThe categorical variables were analysed using the chi-squared test or Fisher's exact test, while the continuous variables were compared using the Mann-Whitney U test. P-values were obtained using bilateral tests, with statistical significance defined as p < 0.05. The statistical analysis was performed with version 22.0 of the SPSS® software (IBM Corp, Chicago, Armonk, NY, USA).
ResultsOf the 632 patients included in the study, 13 (2.1%) had a sample that was not amplified or was invalid for obtaining a consensus sequence using Sanger sequencing. In total, 617 patients with HCV-GT1a were valid for the statistical analysis. Overall, 80.1% (n = 494) of the subjects were male with a mean age of 50 years (47–53). In addition, 52.8% (n = 326) were patients with HCV monoinfection and 47.2% (n = 291) were patients HIV/HCV co-infection. HCV-GT1a clade ii was more heavily represented than clade i (82.5%, n = 509, versus 17.5%, n = 108), and was more prevalent in individuals with HCV monoinfection than HIV/HCV co-infection (87.4% versus 77.0%, p = 0.001).
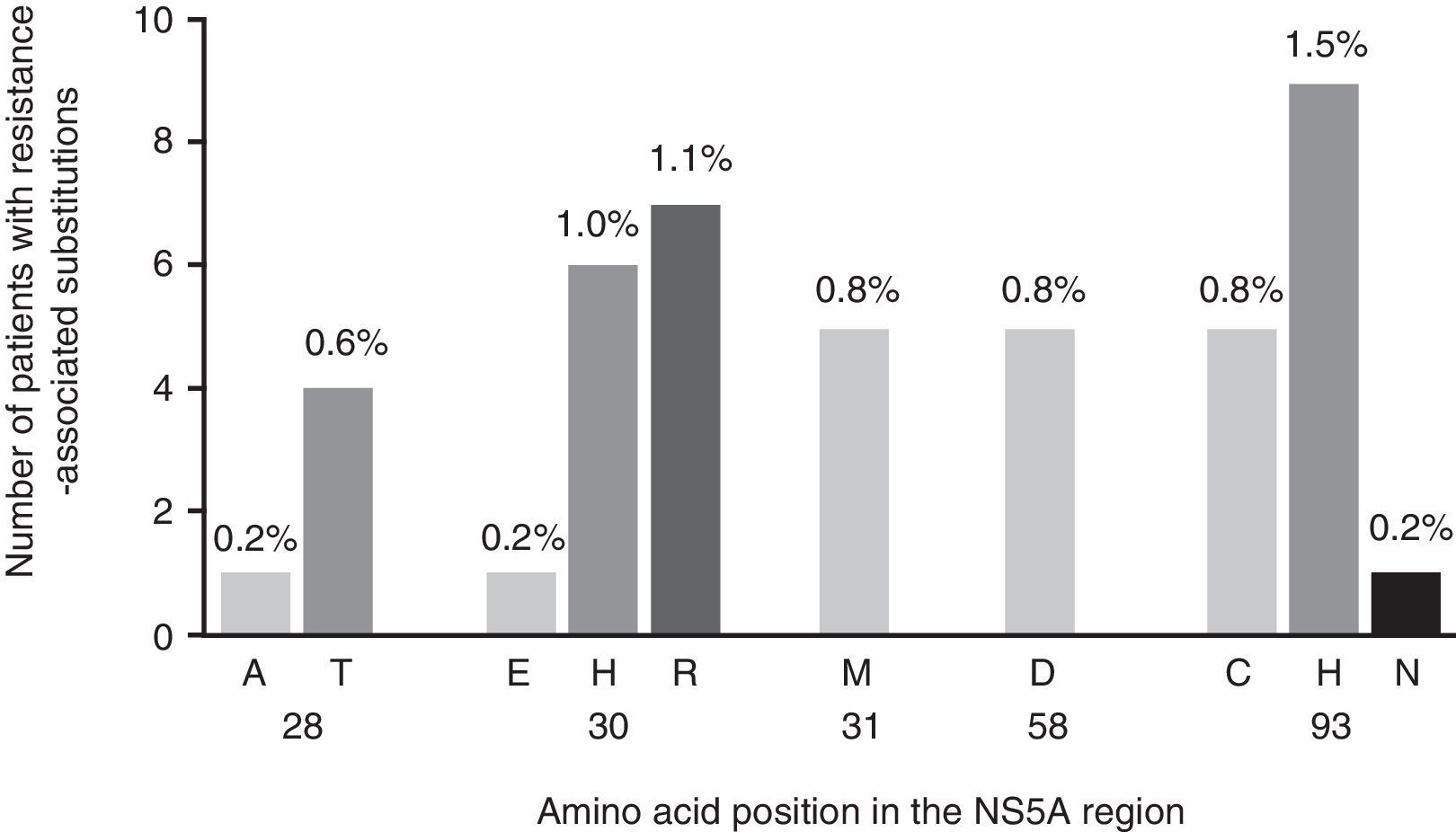
Viruses carrying RASs to EBR were present in 38 samples (6.2%) from patients infected with HCV-GT1a (Fig. 1), 6 of which contained viruses with double RASs. The most common RASs were Y93C/H/N (2.4%; n = 15) and Q30E/H/R (2.3%; n = 14), while M28A/T, L31M and H58D were less heavily represented (0.8%; n = 5 for each). The double mutations Q30H + Y93H and Q30R + Y93H had a low frequency (0.6%, n = 4, and 0.3%, n = 2, respectively) (Fig. 1). Curiously, RASs that conferred reduced sensitivity or susceptibility to EBR were only identified in 21 of the 38 (55.2%) strains identified with RASs to EBR when analysed using g2p[HCV]. This algorithm identified exclusively as principal RASs to EBR the positions Q30H/R (n = 7) and Y93C/H/N (n = 8) as individual mutations and Q30H + Y93H (n = 4) and Q30R + Y93H (n = 2) as double mutations.
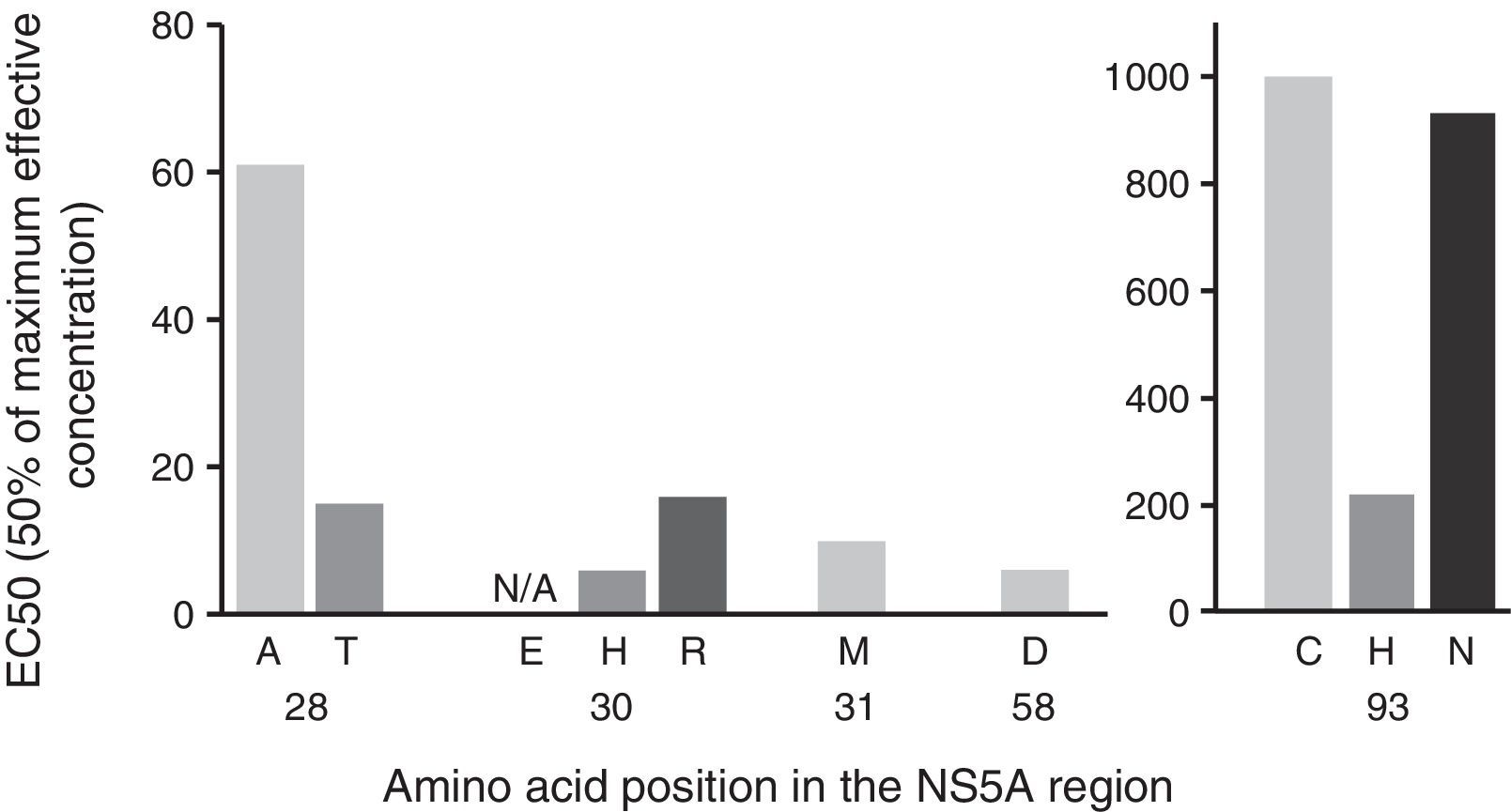
Below, we evaluate the impact of the variants identified based on the change in copy number, with the aim of evaluating the specific weight of each mutation and, therefore, the clinical relevance of the variants found in our cohort (Fig. 2). The mutations identified in the sequences analysed presented a differing pattern of resistance to EBR, varying between 6x and 1235x (Fig. 2). In particular, we identified mutations at the positions Q30H/R and Y93C/H/N as those with the highest level of resistance to EBR (Fig. 2).
Impact of NS5A substitution variants by change in copy number. The complete NS5A gene was sequenced using Sanger sequencing. Pharmacological sensitivity was confirmed using geno2pheno[HCV]. The double mutations Q30H+Y93H and Q30R+Y93H were found in 6 individuals. The analysis of the change in copy number was based on Black et al., 20158 and Cento et al., 2016.14
EC50: concentration necessary to achieve 50% viral replication; NS5A: nonstructural protein 5A.
The influence of the different clades in the presence of RASs to EBR was then evaluated, but no differences were observed (clade i 8.3%, n = 9/108, versus clade ii 5.7%, n = 29/509; p = 0.278). Surprisingly, patients infected with virus who had NS5A RASs conferring reduced susceptibility to EBR were twice as common among those with HIV-HCV co-infection than among those with HCV monoinfection (8.2%, n = 24/291, versus 4.3%, n = 14/326; p = 0.045).
DiscussionIn this study, we observed a low prevalence of NS5A RASs to EBR in HCV-GT1a patients not previously treated with drugs in the NS5A inhibitors family. As far as we know, this is the first study to evaluate the prevalence of NS5A RASs in the HCV-GT1a genotype in Spain. Our data on the prevalence of NS5A RASs to EBR contrast with the 12% recently presented by Zeuzem et al. (2015) in never-treated patients infected with HCV-GT1a involved in the C_EDGE phase 3 study3.
The real-world effect of NS5A RASs to EBR on the response to HCV treatment is not yet clear, and its nature and impact on first-line therapy and treatment strategies continue to present a serious challenge. In addition, the lack of data in clinical practice hinders our ability to compare our results with those from other countries.
Recent studies have brought to light the different distributions of the two clades of HCV-GT1a, which are present in equal proportions in Europe, while clade i is more prevalent in the USA15,16. In our study, we observed a much higher prevalence of clade ii than clade i, with the former representing roughly 4 in every 5 sequences analysed; the latter is significantly less prevalent (17%) than in other European countries (48% for Italian sequences, 53% for German ones). This phylogenetic difference may indicate a possible difference in the temporal propagation of HCV-GT1a clades in Spain in comparison with other European countries, where the two clades are equally distributed. Additionally, in contrast to the data published by De Luca et al. in 2015,15 we observed that clade ii was inversely associated with the presence of HIV infection. The differences in the prevalence of NS5A RASs to EBR by HCV-GT1a clade may offer some explanation of why those variants are less common in European Patients infected with HCV-GT1a than in North American patients, where clade i is more prevalent.15 In our study population, the low prevalence of NS5A RASs to EBR and of HCV-GT1a clade i meant that we were not able to establish any association between the clade and the presence of NS5A RASs. Further studies with larger cohorts will therefore be required in order to clarify the emergence of NS5A RASs to EBR in relation to HCV-GT1a clades.
The latest American Guidelines recommend using resistance tests to identify NS5A RASs in patients infected with HCV-GT1a starting treatment with grazoprevir/EBR.10 In effect, the presence of NS5A RASs and their effect on the response to treatment will influence future therapy choices. In view of the low prevalence observed in our cohort, it may not currently be necessary to perform these tests routinely in Spain. However, it is likely that this scenario will change, and a monitoring system must therefore be implemented that will enable us to evaluate the need to test for NS5A RASs in the near future.
It should be noted that grazoprevir/EBR may be the most cost-effective of the DAAs that have been approved to date, which will probably influence the choice of therapy17. Taken together with the low prevalence of NS5A RASs to EBR observed in our cohort, this will undoubtedly have a bearing on the introduction of grazoprevir/EBR as first-line therapy in Spain.
Some limitations of the study must be taken into account. Firstly, the absence of clinical data has meant that we were unable to evaluate the possible impact of NS5A RASs to EBR on virus response. Nonetheless, clinically relevant RASs were extrapolated from the recently reviewed European Recommendations.5 Secondly, clinical and epidemiological data (for example, mode of transmission of HCV, viral RNA load) were limited and would be needed in order to interpret the results. For example, despite the fact that the Spanish National Health System's most recent strategic plan to combat HCV established general criteria for treatment with DAAs, considering patients with advanced liver fibrosis (F2-F4) and transplant patients or those on the transplant waiting list to be high priority,18 we cannot guarantee that all patients included in the study had advanced liver fibrosis. In spite of this, it should be noted that our study uses a national sample that is representative of the entire population living with chronic HCV infection in Spain. This may help us to have a global view of the current situation in the Spanish population, in contrast to studies conducted in individual hospitals. Thirdly, Sanger sequencing (HCV population sequencing) was used instead of massively parallel sequencing to identify NS5A RASs. Although Sanger sequencing may slightly underestimate the prevalence of NS5A RASs to EBR, the impact of RASs on the efficacy of grazoprevir/EBR appears to disappear after administering EBR/grazoprevir + RBV for 16 weeks.6 In addition, the need to quickly diagnose NS5A RASs is an argument against massively parallel sequencing due to its greater response time and cost.
In conclusion, our data show that, in Spain, among patients infected with HCV-GT1a not previously treated with NS5A inhibitors, there is a low prevalence of NS5A RASs to EBR. This information could be essential in guiding the implementation of this DAA in Spain, and in managing patients infected with HCV-GT1a in the near future.
FundingThis study was funded by the Instituto de Salud Carlos III (Ref. PI14CIII/00011 to SR, MPY 1039/14 and PI15CIII/00031 to VB). CP is supported by the Portuguese Fundação para a Ciência e Tecnologia (grant number SFRH/BPD/77448/2011, part of the EDCTP2 programme funded by the European Union). VB is funded by the Miguel Servet programme directed by the Fondo de Investigación Sanitaria [Health Research Fund] (Instituto de Salud Carlos III) (grant number CP13/00098).
AuthorshipVB conceived and designed the study; MSC, BEC and VB obtained the data, CP, IMC and VB analysed and interpreted the data; CP, BEC and VB prepared the manuscript; SR and IMC performed a critical review of the article; CP, SR and VB approved the final version. SR and VB contributed equally.
Conflicts of interestThe authors declare that they have no conflicts of interest.
The authors would like to thank for their participation: all of the patients, the collaborating centres, and the clinical samples provided.
The following individuals and institutions participated in this epidemiological survey:
- 1.
Área Sanitaria de Ferrol: P. Ordoñez Barrosa.
- 2.
Centro Médico de Asturias: R. Vilches Vilches.
- 3.
Complejo Asistencial Universitario de Burgos: Gonzalo F. Saez-Royuela.
- 4.
Complejo Hospitalario de Navarra: I. Polo Vigas.
- 5.
Complejo Hospitalario de Orense: G. Esteban Meruéndano.
- 6.
Complejo Hospitalario de Pontevedra: M. Trigo Daporta.
- 7.
Complejo Hospitalario Universitario de Santiago: A. Aguilera Guirao.
- 8.
Complejo Hospitalario Universitario La Coruña: M. A. Cañizares Castellanos.
- 9.
Complejo Hospitalario Xeral-Calde: A. Coira Nieto.
- 10.
Fundación Hospital Alcorcón: M. L. Casas Losada.
- 11.
Fundación Hospital de Jove: E. Hidalgo Pérez.
- 12.
Fundación Jiménez Díaz-Ute: R. Fernández Roblas.
- 13.
Gerencia del Área de Salud de Badajoz, Llerena y Zafra: G. Sánchez Alor.
- 14.
Gestión Sanitaria de Mallorca (GESMA): V. Fernández Baca Gutiérrez del Álamo.
- 15.
Hospital Arnau de Vilanova de Valencia: R. Giner Duran.
- 16.
Hospital Can Misses: A. Hurtado Fernandez.
- 17.
Hospital Central de Asturias: S. Melón Garcia.
- 18.
Hospital Central de la Defensa Gómez Ulla: M. Mateo Maestre.
- 19.
Hospital Clínic i Provincial de Barcelona: F. Marco Reverte.
- 20.
Hospital Clínico San Carlos: E. Culebras Lopez.
- 21.
Hospital Clínico Universitario de Salamanca: M. N. Gutierrez Zufiaurre.
- 22.
Hospital Clínico Universitario de Valladolid: C. Hinojosa Mena Bernal.
- 23.
Hospital Clínico Universitario Lozano Blesa: C. Rubio Calvo.
- 24.
Hospital Comarcal de Inca: J. Saurina Gomila.
- 25.
Hospital de Basurto: S. Hernaez Crespo.
- 26.
Hospital de Cabueñes: L. Otero Guerra.
- 27.
Hospital de Cruces: L. López Soria.
- 28.
Hospital de Donostia: C.G. Cilla Eguiluz.
- 29.
Hospital de Especialidades de Jerez de la Frontera: M. D. López Prieto.
- 30.
Hospital de Especialidades de Puerto Real: I. Jesús de la Calle.
- 31.
Hospital de Gran Canaria Dr. Negrín: M. J. Pena López.
- 32.
Hospital de Hellín: C. Romero Portilla.
- 33.
Hospital de la Agencia Valenciana de Salud Vega Baja: N. Gonzalo Jiménez.
- 34.
Hospital de la Línea de la Concepción: F. J. Casas Ciria.
- 35.
Hospital de Móstoles: F. López Fabal.
- 36.
Hospital de Palamós: N. Guinart Sola.
- 37.
Hospital de Poniente: T. Cabezas Fernández.
- 38.
Hospital do Meixoeiro: S. Pérez Castro.
- 39.
Hospital Don Benito-Villanueva de la Serena: A. Valle Valencia.
- 40.
Hospital El Bierzo: C. Raya Fernández.
- 41.
Hospital Ernest Lluch Martin: B. Fortuño Cebamanos.
- 42.
Hospital Galdakao-Usansolo: M. J. Lopez de Goicoechea San Román.
- 43.
Hospital García Orcoyen: L. Barrado Blanco.
- 44.
Hospital General de Castellón: B. Gomila Sard.
- 45.
Hospital General de Lanzarote: R. Copado Carretero.
- 46.
Hospital General de Segovia: J. Elizaga Corrales.
- 47.
Hospital General Juan Ramón Jiménez: J. Saavedra Martin.
- 48.
Hospital General Río Carrión: T. García Valero.
- 49.
Hospital General San Jorge: L. Torres Sopena.
- 50.
Hospital General Universitario de Alicante: A. Gimeno Gascón.
- 51.
Hospital General Universitario de Elche: M. Ruiz García.
- 52.
Hospital General Universitario Gregorio Marañón: T. VicenteRangel.
- 53.
Hospital General Universitario Reina Sofía: A. J. Marín Cervantes.
- 54.
Hospital Infanta Cristina (Badajoz): R. Sánchez Silos.
- 55.
Hospital Infanta Elena: M. de la Iglesia Salgado.
- 56.
Hospital J. M. Morales Meseguer: C. Guerrero Gómez.
- 57.
Hospital Mateu Orfila: L. Carbo.
- 58.
Hospital Nuestra Señora de Sonsoles: A. Gómez del Campo Dechado.
- 59.
Hospital Povisa S. A.: M. D. Martínez Otero.
- 60.
Hospital Puerta de Hierro (Majadahonda): F. Portero Azorin.
- 61.
Hospital Rafael Méndez: E. Cascales Alcolea.
- 62.
Hospital Reina Sofía (Córdoba): F. Rodríguez Cantalejo.
- 63.
Hospital Reina Sofía (Tudela): J. J. García Irure.
- 64.
Hospital San Agustín (Asturias): G. Sierra Dorado.
- 65.
Hospital San Pedro: C. Martínez Gil.
- 66.
Hospital Santa Maria Nai: J. Garcia Costa.
- 67.
Hospital Txagorritxu: M. J. Lezaun Bugui.
- 68.
Hospital Universitario Marqués de Valdecilla: J. Crespo García.
- 69.
Hospital Universitario de Canarias: F. Díaz-Flores Estevez.
- 70.
Hospital Universitario de Ceuta: J. López Barba.
- 71.
Hospital Universitario de La Princesa: L. Cardeñoso Domingo.
- 72.
Hospital Universitario Dr. Peset: J. Alberola Enguidanos.
- 73.
Hospital Universitario Insular de Gran Canaria: E. Santana Rodriguez.
- 74.
Hospital Universitario Miguel Servet: A. Martínez Sapiña.
- 75.
Hospital V. Alvarez Buylla: M. C. Galarraga Gay.
- 76.
Hospital Virgen de la Concha: R. Martínez González.
- 77.
Hospital Virgen de la Salud (Toledo): P. Zamarrón Fuertes.
- 78.
Hospital Virgen de la Victoria: E. Clavijo Frutos.
- 79.
Hospital Virgen del Castillo: M. L. López Yepes.
- 80.
Hospital Virgen del Puerto: C. García Tejero.
- 81.
Hospital Virgen del Rocío: L. Merino Díaz.
- 82.
Laboratorio de Referencia del Camp de Tarragona i Terres del’Ebre: M. J. Puerta Martínez.
- 83.
Laboratorio de Referencia de Catalunya (El Prat de Llobregat): M. Salvado Costa M; G. Soria Guerrero.
- 84.
Laboratorio BR Salud: E. Aznar Cano.
Please cite this article as: Palladino C, Esteban-Cartelle B, Mate-Cano I, Sánchez-Carrillo M, Resino S, Briz V. Frecuencia de sustituciones relevantes asociadas a resistencia en la región NS5A a elbasvir en el virus de la hepatitis C en pacientes con genotipo 1a en España. Enferm Infecc Microbiol Clin. 2018;36:262–267.






![Impact of NS5A substitution variants by change in copy number. The complete NS5A gene was sequenced using Sanger sequencing. Pharmacological sensitivity was confirmed using geno2pheno[HCV]. The double mutations Q30H+Y93H and Q30R+Y93H were found in 6 individuals. The analysis of the change in copy number was based on Black et al., 20158 and Cento et al., 2016.14 EC50: concentration necessary to achieve 50% viral replication; NS5A: nonstructural protein 5A. Impact of NS5A substitution variants by change in copy number. The complete NS5A gene was sequenced using Sanger sequencing. Pharmacological sensitivity was confirmed using geno2pheno[HCV]. The double mutations Q30H+Y93H and Q30R+Y93H were found in 6 individuals. The analysis of the change in copy number was based on Black et al., 20158 and Cento et al., 2016.14 EC50: concentration necessary to achieve 50% viral replication; NS5A: nonstructural protein 5A.](https://static.elsevier.es/multimedia/2529993X/0000003600000005/v1_201805030412/S2529993X18300868/v1_201805030412/en/main.assets/thumbnail/gr2.jpeg?xkr=ue/ImdikoIMrsJoerZ+w96p5LBcBpyJTqfwgorxm+Ow=)
