Here, we propose a novel modified Carba NP test for detecting KPC-producing Enterobacterales using imipenem/relebactam.
Material and methodsThe test performance was evaluated in a random selection of 160 previously molecularly characterized clinical isolates carrying the 110 blaKPC, 1 blaGES, 12 blaVIM, 4 blaIMP, 3 blaNDM and 42 blaOXA-48-like genes. The proposed method relies on the detection of imipenem hydrolysis in an imipenem/relebactam antibiotic solution and subsequent visual interpretation by color change.
ResultsAll class A producing Enterobacterales (111/111) were detected using imipenem/relebactam as no visual appreciation of color change was perceived due to a nule hydrolysis of imipenem in the antibiotic solution. Overall, the assay showed 100% sensitivity (111/111) and specificity (69/69) for detecting class A KPC-producing Enterobacterales.
DiscussionThe biochemical assay provides very reliable results for detecting KPC-producing Enterobacterales, with a turnaround time of less than 1 hour, minimum handling and no specialized equipment required.
En este manuscrito proponemos un novedoso ensayo basado en el Carba NP test para la detección de Enterobacterales productoras de KPC utilizando imipenem/relebactam.
Material y métodosLa evaluación se realizó en una selección aleatoria de 160 aislados clínicos previamente caracterizados molecularmente que portaban los genes 110 blaKPC, 1 blaGES, 12 blaVIM, 4 blaIMP, 3 blaNDM y 42 blaOXA-48-like. El método propuesto se basa en la detección de la hidrólisis de imipenem en una solución de imipenem/relebactam y posteriormente su interpretación mediante un cambio de color.
ResultadosEl ensayo tiene una sensibilidad (111/111) y una especificidad (69/69) del 100% para detectar Enterobacterales productoras de KPC, de clase A.
DiscusiónEste ensayo bioquímico proporciona resultados muy fiables para la detección de Enterobacterales productoras de KPC, con un tiempo de respuesta inferior a 1 h, además de una manipulación mínima y sin necesidad de ningún equipamiento especializado.
The global rise in the incidence of carbapenemase-producing Enterobacterales (CPE) is alarming and poses a challenge to health services worldwide.1 This is especially worrying in relation to Klebsiella pneumoniae carbapenemase (KPC)-producing bacteria, the most frequently occurring type of CPE worldwide.2 Limited therapeutic options are available for CPE, and only “second line” drugs such as polymyxins, tigecycline, aminoglycosides and fosfomycin are generally used. Double carbapenem therapy can also be considered, as can combination therapies, which are associated with better outcomes for high-risk patients.3 Novel β-lactam inhibitors are being developed with the aim of restoring the activity of β-lactam antibiotics against CPE. Relebactam (MSD, USA) inhibits class A and C β-lactamases and is currently under clinical development in combination with imipenem-cilastatin. Imipenem/relebactam has recently undergone testing in phase 3 clinical trials to treat patients with complicated intra-abdominal infections, complicated urinary tract infections and hospital-acquired/ventilator-associated bacterial pneumonia, with promising results. Biochemical methods have been used to detect carbapenem hydrolysis as a surrogate test for susceptibility.4–7 EUCAST guidelines for carbapenemase detection also recommend biochemical assays as useful methods in clinical practice. These methods measure the color change in the medium due to the presence of a pH indicator, which shifts when the carbapenem antibiotic is hydrolysed as a consequence of acidification. Here, we propose a modified Carba NP test for detecting class A KPC-producing Enterobacterales. We focused on KPC-producing isolates (because of their high prevalence) and used imipenem/relebactam as the reaction substrate. This approach makes use of the capacity of relebactam to inhibit imipenem hydrolysis and thus confirm the presence of class A carbapenemase, the only class inhibited by this compound.
Material and methodsBacterial isolatesThe proposed method was applied to a random selection of 172 carbapenemase-producing clinical isolates previously molecularly characterized using the Xpert Carba-R Assay (Cepheid, Sunnyvale, USA). The isolates carried 110 blaKPC, 1 blaGES, 12 blaVIM, 4 blaIMP, 3 blaNDM and 42 blaOXA-48-like genes.8 In the case of the GES-producing isolate, WGS was performed to determine the underlying mechanism of resistance. The molecular results were used as gold standard. As negative controls clinical isolates, Enterobacter cloacae (n=2), Citrobacter freundii (n=2), and Serratia marcescens (n=2), were used. Also, reference strains as K. pneumoniae ATCC 700613 and E. coli ATCC 35218 were tested. Species identification was confirmed by MALDI-TOF MS. The CPE comprised 120 K. pneumoniae, 3 Klebsiella oxytoca, 12 Escherichia coli, 18 Enterobacter cloacae, 17 Citrobacter freundii and 2 Serratia marcescens. E. coli ATCC 25922 was used as a negative control, and a PCR-confirmed KPC-producing E. coli was used as a positive control in every assay.
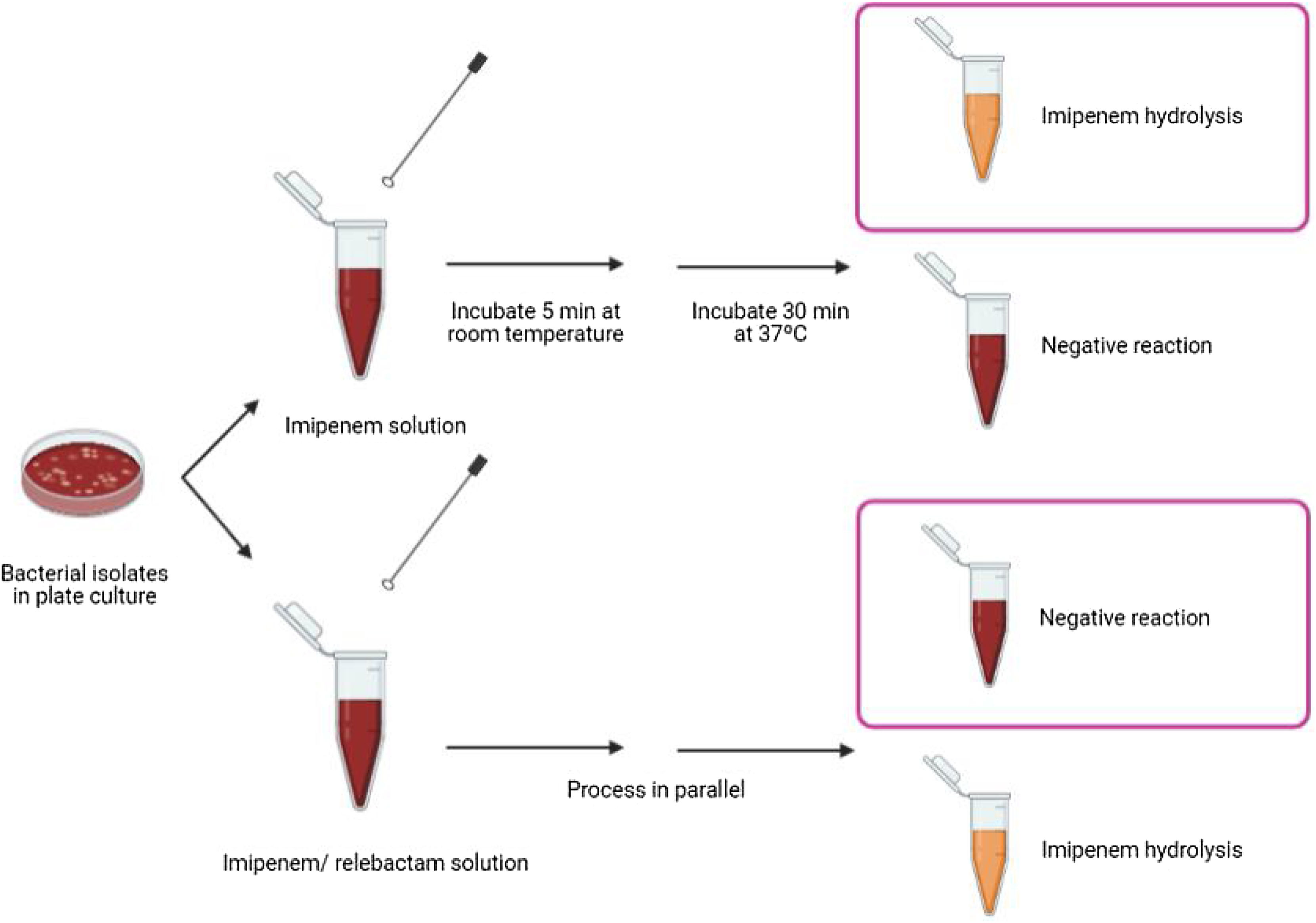
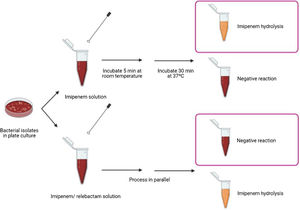
Biochemical assayThe proposed method relies on the detection of imipenem hydrolysis in an imipenem/relebactam antibiotic combination by using a modified Carba NP test,7 a buffer developed by the authors of this paper9 and subsequent visual interpretation of the results. The test was performed as follows: 1μl loop of the different bacterial isolates were resuspended in a tube containing 100μl of imipenem (3mg/ml) dissolved in 0.5% phenol red solution, 10mM NH4HCO3, 10mM ZnSO4 and 1% of SDS (pH=8) and in a second tube containing imipenem/relebactam (3mg/ml/4mg/ml) dissolved in the same buffer. Imipenem hydrolysis was also measured as an internal control. The suspensions were incubated at room temperature for 5min and then incubated at 37°C under agitation for another 30min. The test results were interpreted by technicians who were blinded to sample identification. Class A carbapenemases were identified when the imipenem solution turned yellow or orange-yellow and the imipenem/relebactam solution remained red, due to the impossibility of the enzyme to hydrolyze imipenem in the presence of relebactam (Fig. 1).
Class A carbapenemases detection in Enterobacterales by using a modified Carba NP test with imipenem/relebactam. Representative results obtained using a modified Carba NP test with imipenem/relebactam. Class A carbapenemases hydrolyze imipenem providing a color change in the imipenem solution whereas they are incapable of hydrolysing imipenem when incubated with relebactam, providing no color change in the antibiotic solution.
After incubation of the bacterial isolates with imipenem for 30min at 37°C, we observed a color change from red to yellow in all carbapenemase-producing isolates. The test showed 100% sensitivity and specificity for detecting carbapenemase activity, as assessed by comparison with molecular-based methods. Regarding imipenem hydrolysis in the class A carbapenemases (n=111), the color change was obvious after incubation for 15min at room temperature, but was clear as early as after 5min for most isolates (n=103). In the imipenem/relebactam combination, all isolates yielded a red color, irrespective of the incubation time, demonstrating the inhibitory capacity of relebactam and the susceptibility of the isolates to the imipenem/relebactam combination. Regarding the class B carbapenemases (n=19), for most isolates (n=10) the color change in the tube containing imipenem was clear after incubation for 5min at room temperature and very obvious after incubation for 15min at 37°C. For class D carbapenemases (n=42), the color change in the tube containing imipenem was not as evident after incubation for 30min at 37°C. None of the isolates belonging to class B and D carbapenemases yielded a red color in the tube containing imipenem/relebactam after incubation for 30min, demonstrating the inability of relebactam to inhibit these carbapenemases. These results are consistent with those of the molecular methods, yielding 100% sensitivity (111/111) and specificity (69/69) for detection of class A producing Enterobacterales susceptible to imipenem/relebactam.
Carbapenemase non-producing isolates did not hydrolyze imipenem, as demonstrated by the absence of color change in the imipenem solution, thus in the imipenem/relebactam.
DiscussionWe report here a novel approach for the rapid detection of class A carbapenemases using a modified Carba NP test with imipenem/relebactam as a substrate for the hydrolysis reaction. The study findings support previous data concerning the use of modified Carba NP tests for detecting carbapenemase activity in Enterobacterales.4–7
There are currently few or no therapeutic options available for treating infections caused by KPC-producing Enterobacterales, which are usually multidrug resistant, especially since the emergence of ceftazidime-avibactam resistance.10–11 The early and accurate detection of these isolates is therefore extremely important in regard to prescribing targeted treatments.
The study findings showed that this novel Carba-NP-based method is an excellent tool for detecting KPC-producing Enterobacterales, providing results with a turnaround time of less than 1h. The method yielded 100% sensitivity and specificity (111/111; 69/69), as indicated by comparison with the molecular characterization of the isolates. The findings also revealed that the incubation buffer strongly affected imipenem hydrolysis. Low pH incubation buffers (e.g. NH4-citrate, pH 6) have yielded false-negative results in isolates with OXA-type enzymes.9,12 Conversely, in the present study the isolates showed high reactivity with NH4HCO3 buffer (pH 8) and shorter reaction times. Inclusion of SDS in the incubation buffer facilitated the reactivity in hypermucoviscous K. pneumoniae.
In addition to the KPC-producing isolates, we tested one GES-producing K. oxytoca, in which the negative hydrolysis of imipenem/relebactam appeared consistent with the results obtained with the KPC-producing isolates. However, further isolates must be tested to confirm the results. The biochemical assay exclusively detects the presence of carbapenemase enzymes, but does not detect carbapenem resistance due to other resistance mechanisms such as OmpK35 disruption and/or mutated OmpK36, which have been described as chromosomal resistance in these isolates.13
This in-house method is rapid, simple and inexpensive, making it very useful for the identification of KPC-producing Enterobacterales in clinical microbiological laboratories. The rapid identification of these multidrug resistant bacteria will help in the early administration of targeted treatments such as imipenem/relebactam, which have produced excellent outcomes.14,15 Other commercial and in-house developed methods have a turnaround time of 2h, but the proposed modified Carba-NP test provides results in less than 1h, with minimum handling and no need for specialized equipment. The method could therefore be easily implemented in microbiologial laboratories, even in low-resource settings.
FundingThe results of this work have been funded by the Projects N° PI15/00860 and PI18/00501, integrated in the National Plan for Scientific Research, Development and Technological Innovation 2013–2016 and funded by the ISCIII-General Subdirection of Assessment and Promotion of the Research – European Regional Development Fund (FEDER) “A way of making Europe”.
Marina Oviaño is supported by the Juan Rodés Program (JR 18/00006) from the Health Research Fund (FIS) of the ISCIII.
Germán Bou was in receipt of a grant from the Investigator Initiated Studies Program, funded by Merck Sharp and Dohme (MSD). The company declare not having of competence in the design and results of the study.
Conflict of interestsThe authors declare having no conflict of interest.