We evaluated the presence of sIgA in saliva, versus Escherichia coli secreted proteins (Esp) related to the type III secretion system (T3SS), and its semi-quantitative concentration in children under 2 years-old (no longer breastfed) who were previously colonized or infected with enteropathogenic E. coli (EPEC).
MethodsWe analyzed the presence of sIgA in 40 children, who previously had positive cultures for EPEC associated (n=17) or not associated (n=23) with diarrhea, using the Western Blot technique versus E. coli secreted proteins: EspABCD. A semi-quantitative measurement of the reaction for each protein was made by its density peaks (OD).
ResultsWe found sIgA versus all or some EspABCD proteins in both groups. However, the ill patients had higher concentrations of these antibodies than colonized patients.
DiscussionThe presence of sIgA in saliva could reflect an intestinal immune response and their levels could be related to a greater exposure and/or bacterial load.
Evaluamos la presencia de inmunoglobulina secretoria A (sIgA) frente a proteínas secretadas por E. coli (Esp) relacionadas con el sistema de secreción tipo III (T3SS) en saliva, y su concentración semicuantitativa en niños menores de 2 años (no lactantes) colonizados o infectados previamente con E. coli enteropatógena (EPEC).
MétodosAnalizamos la presencia de sIgA en 40 niños con cultivos positivos previos para EPEC asociados (n=17) o no (n=23) con diarrea, mediante la técnica de Western-blot frente a las proteínas secretadas por E. coli (EspABCD), realizando mediciones semicuantitativas de la reacción de cada proteína mediante sus picos de densidad (OD).
ResultadosAmbos grupos presentaron sIgA frente a las proteínas EspABCD, aunque los pacientes enfermos presentaron mayores concentraciones de estos anticuerpos que los colonizados.
DiscusiónLa presencia de sIgA en la saliva podría reflejar una respuesta inmune intestinal y sus niveles podrían estar relacionados con una mayor exposición y/o carga bacteriana.
Diarrhea continues to be the second most common cause of death in children less than 5 years of age worldwide, with enteropathogenic Escherichiacoli (EPEC) as one of the most frequent etiological agents.1,2 These pathogens bind to the enterocytes producing an attaching and effacing lesion in intestinal epithelial cells, mediated by a type III secretion system (T3SS), which forms a structure known as injectosome/needle-complex. Some translocated effectors are called E. coli secreted proteins (Esp), such as EspA multimers form the extension of the injectosome, while EspB and EspD form a pore in the host-cell membrane that enables entry of additional effector proteins into the cytoplasm of the enterocyte.1 EspC is an autotransporter protein secreted by a type V secretion system, but can also be secreted through the T3SS and is associated with apoptosis and necrosis.1,3
Antibodies produced in the intestinal mucosa, mainly secretory immunoglobulin A (sIgA), are thought to be responsible for preventing the colonization and adhesion of bacteria to the enterocytes. The production of sIgA in the stroma of the salivary gland is due mostly to antigen-sensitized lymphocytes in the gut-associated lymphoid tissue (GALT) that migrate via the bloodstream to mucosal tissues to form the mucosa-associated lymphoid tissue (MALT). At these extraintestinal sites, B-cells differentiate into plasma cells and produce antigen-specific secretory IgA.4 Since the collection of saliva is a simple, rapid, and non-invasive method, we conducted this study to determine the presence, and to compare the amount of sIgA in saliva against specific virulence proteins of EPEC (EspABCD) in children under 2 years-old who were previously colonized (asymptomatic) or infected with EPEC.
MethodsWe studied 40 children between 12 and 24 months of age who were no longer breastfeeded and who previously had positive cultures for EPEC associated (n=17) or non-associated (n=23) with diarrhea. These children were part of an active surveillance diarrhea cohort study in peri-urban communities in Lima, Peru.5 Diarrhea samples were collected at the beginning of an episode (more than 3 loose stools per day or at least one loose stool with blood) and control samples were considered as colonization if there were no loose stools one week before and after sample collection. Detection of EPEC was done by determining the presence of the eae gene using a real-time multiplex PCR for all six groups of diarrheagenic E. coli.6 Participating parents signed an informed consent prior to data and sample collection. The study was approved by the Institutional Review Board of Universidad Peruana Cayetano Heredia in Lima, Peru.
From each child we collected 500μL of un-stimulated whole saliva samples from the floor of the mouth using sterile polypropylene graduated transfer pipettes. The sample collection was performed at least 1 hour after feeding to avoid contamination with non-salivary components. Saliva samples were centrifuged according to previously described methods.7
To study the Esp-specific sIgA, we used the EPEC prototype strain E2348/69. Since EspB and EspD proteins have similar molecular weights, we used two mutant strains: E2348/69ΔespB and E2348/69ΔespD, that express either EspD or EspB, respectively. To induce the secretion of virulence proteins and their extraction from culture supernatants, the strains were grown and treated as described previously.8 Through a vertical denaturing electrophoresis (SDS-PAGE: 12.5%) and stained with Coomassie Blue we determined the secretion profiles of EPEC. Western blots were then done using a 1/10 dilution for the primary antibody (sIgA Saliva), 1/200 for the secondary antibody (anti-human anti-IgA conjugated) and diaminobenzidine (DAB) for development. The presence of each protein was determined based on the molecular weight previously reported. The intensity of each Western Blot bands were analyzed using the Quantity One v4.6.3 software (Bio-Rad, Hercules, CA, USA), which measures the band intensity by OD (peaks density). Boxplot and analysis of variance (ANOVA) were used to compare the intensity for each protein in ill and colonized infants.
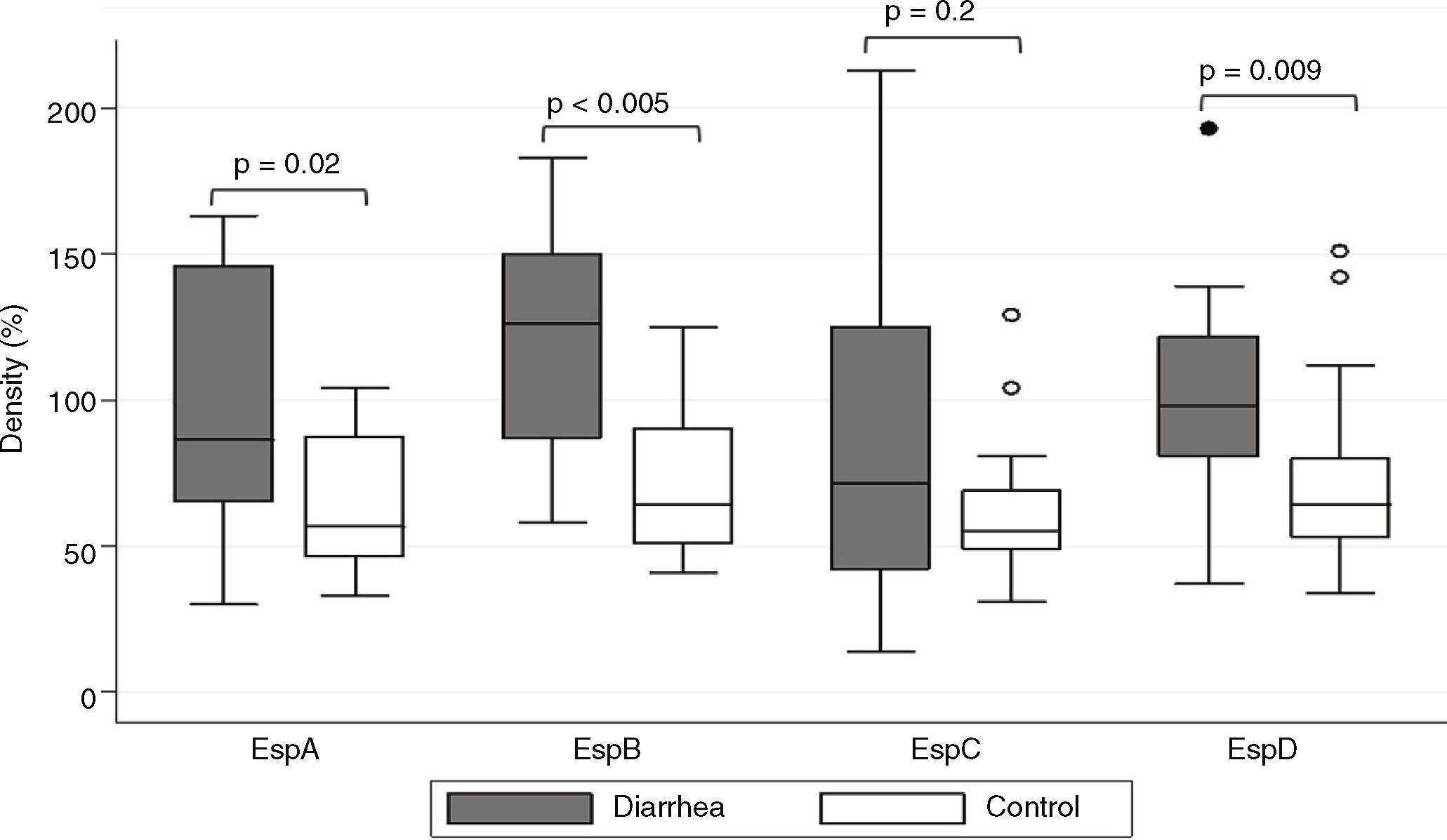
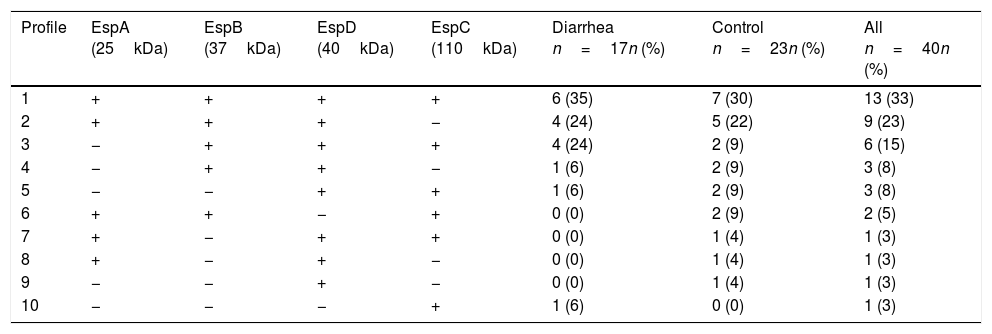
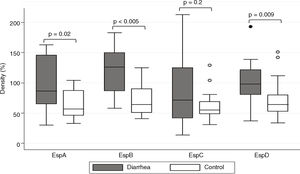
ResultsSaliva samples were taken 8–90 days after EPEC detection. The presence of Esp-specific sIgA in saliva is presented in relation to the age, the type of EPEC infection (diarrhea or control), and the time between the positive EPEC stool samples and the saliva collection in supplementary Table 1. Of interest, 3 children had a documented previous EPEC infection and 2 children had a documented previous Shiga-toxin producing E. coli (STEC) infection in the second year of life, in addition to the current EPEC infection. The most frequent Esp-specific sIgA was EspD in 37 samples (93%), followed by EspB in 33 samples (83%), EspA and EspC, both with 26 samples (65%). There were no significant differences (p<0.05) in the detection of Esp-specific sIgA between diarrhea and control groups, respectively: EspA: 10/17 (59%) vs. 16/23 (70%); EspB: 15/17 (88%) vs. 18/23 (78%); EspC: 12/17 (71%) vs. 14/23 (61%); and EspD: 16/17 (94%) vs. 21/23 (91%). However; when we compared band intensities for each protein in both groups, there were statistically significant differences for each one, except for EspC (Fig. 1). The most common combination of antibodies was the presence of sIgA for all 4 proteins in 33%, followed by EspA/EspB/EspD in 23%, and EspB/EspC/EspD in 15% of samples. Ten antibodies’ profiles were obtained (Table 1). The high frequency of antibodies against EPEC Esp-proteins found in this population may not only represent the memory response to the last documented EPEC infection, but may also represent the immunological response to multiple previous infections or colonizations with EPEC since birth.
Profile of sIgA antibodies in saliva against the Esp-proteins.
| Profile | EspA (25kDa) | EspB (37kDa) | EspD (40kDa) | EspC (110kDa) | Diarrhea n=17n (%) | Control n=23n (%) | All n=40n (%) |
|---|---|---|---|---|---|---|---|
| 1 | + | + | + | + | 6 (35) | 7 (30) | 13 (33) |
| 2 | + | + | + | − | 4 (24) | 5 (22) | 9 (23) |
| 3 | − | + | + | + | 4 (24) | 2 (9) | 6 (15) |
| 4 | − | + | + | − | 1 (6) | 2 (9) | 3 (8) |
| 5 | − | − | + | + | 1 (6) | 2 (9) | 3 (8) |
| 6 | + | + | − | + | 0 (0) | 2 (9) | 2 (5) |
| 7 | + | − | + | + | 0 (0) | 1 (4) | 1 (3) |
| 8 | + | − | + | − | 0 (0) | 1 (4) | 1 (3) |
| 9 | − | − | + | − | 0 (0) | 1 (4) | 1 (3) |
| 10 | − | − | − | + | 1 (6) | 0 (0) | 1 (3) |
A previous study in Brazilian children younger than 2 years of age found a high prevalence of EPEC antibodies against intimin, Tir, EspA, EspB and Bfp in serum and saliva samples.9 The detection of antibodies occurred as early as 2 months of age, representing an early exposure to these pathogens in endemic areas, when the frequency of antibody detection in most samples increased with age. Of interest, in serum samples, the most common antibody, anti-EspB, was present in all samples in all age groups. Other studies have evaluated the presence of antibodies against the same proteins of EPEC in colostrum samples from mothers and serum samples from children in these settings.8,10,11
Previous studies on sIgA in saliva against other diarrheagenic E. coli have focused on the detection of antibodies against colonization factor antigen 1 (CFA/1) of enterotoxigenic E. coli (ETEC),11 and against LPS of E. coli O157 in children with hemolytic uremic syndrome (HUS) infected with STEC and healthy controls.7 Of interest, some children with STEC isolates belonging to other O-groups (O26, O145 and O165) lacked detectable O157 LPS-specific antibodies.
EPEC specific O-serotypes did not have correlation with their EPEC strains isolated12 and are not informative to measure antibodies against O-serotypes. EPEC, which is defined by the presence of the intimin gene (eae), belongs to many serogroups,1 therefore, the detection of antibodies against surface-expressed virulence proteins of the T3SS is LPS-independent and universal. Such antibodies provide a clearer picture of immune responses that are EPEC specific, although the rare STEC infection can also induce such antibodies.
We found high band intensities in proteins associated to T3SS in the diarrhea group (p<0.05). These levels of sIgA in saliva may still reflect an intestinal immune response after stimulation for the presence of an enteric pathogen due to the migration of B-cell from the gut to the salivary gland4; immunological memory cells can maintain the response for a long-time.13 The higher levels of specific sIgA could be related with an increase of EPEC load during diarrheal episodes compared with colonized children.14
In summary, we found the presence of anti-Esps antibodies in saliva in both groups, with higher quantity in ill compared to colonized infants. It is necessary to study IgA not only as the first-line of immunological protection against pathogens/toxins, but also to study its effect on organization of the intestinal microbiota, and regulation of intestinal host–microorganism homeostasis.
The study of these antibodies may be useful in better defining first infections and in assessing the role of these antibodies in symptoms severity during subsequent infections. The timing of specific antibody development, and perhaps the level of specific antibodies associated with symptoms, may be a critical variable to address in future studies. Future cohort studies are needed to determine how early children in endemic areas develop an immune response to this group of pathogens, and to define the role of previous infections (symptomatic or asymptomatic) in modulation of the immune response.
FundingThis work was supported by the Public Health Service award 1K01TW007405 (T.J.O.) from the National Institutes of Health, USA; the Agencia Española de Cooperación Internacional para el Desarrollo (AECID) [grant nos. D/019499/08, D/024648/09, D/030509/10, and A1/035720/11]. JR had a fellowship from the program I3 of the Instituto de Salud Carlos III (ISCIII, Spain) [grant no. CES11/012] and ISGlobal is a member of the CERCA Programme, Generalitat de Catalunya.
Conflict of interestsThe authors declare no conflict of interests.
We will like to thank Dr. Jose Luis Puente, from the Department of Molecular Microbiology, UNAM, Mexico, for kindly providing the two mutant strains: E2348/69ΔespB and E2348/69ΔespD.