Wheat is a common cereal in the Western diet and an important source of protein as well as fiber. However, some individuals develop adverse reactions to a wheat-containing diet. The best characterized is celiac disease which develops after intake of gluten in individuals with genetic predisposition. Other wheat-related conditions are less well defined in terms of diagnosis, specific trigger and underlying pathways. Despite this, the overall prevalence of wheat-related disorders has increased in the last decades and the role of microbial factors has been suggested. Several studies have described an altered intestinal microbiota in celiac patients compared to healthy subjects, but less information is available regarding other wheat-related disorders. Here, we discuss the importance of the intestinal microbiota in the metabolism of wheat proteins and the development of inflammatory or functional conditions. Understanding these interactions will open new directions for therapeutic development using bacteria with optimal wheat protein degrading capacity.
El trigo es un cereal frecuente en la dieta occidental y una importante fuente de proteínas y fibra. Sin embargo, algunas personas presentan reacciones adversas a una dieta con trigo. La más conocida es la enfermedad celíaca, que se manifiesta después del consumo de gluten por parte de individuos con predisposición genética. Otras enfermedades relacionadas con el trigo no están tan bien definidas por lo que respecta al diagnóstico, el desencadenante específico y las vías subyacentes. A pesar de ello, la incidencia general de trastornos relacionados con el trigo ha aumentado en las últimas décadas, y se ha sugerido el papel de factores microbianos. Varios estudios han descrito cambios de la microbiota intestinal en pacientes celíacos frente a individuos sanos, pero hay menos información sobre otros trastornos relacionados con el trigo. En este artículo tratamos la importancia de la microbiota intestinal en el metabolismo de las proteínas del trigo y el desarrollo de trastornos inflamatorios o funcionales. El conocimiento de estas interacciones abrirá nuevas vías para el desarrollo terapéutico con bacterias con una capacidad óptima de degradación de las proteínas del trigo.
Cereals are the most important crops worldwide with about 2000 million tons of grain per year.1 The FAO‘s (Food and Agriculture Organization of the United Nations) latest forecast for global cereal production in 2018 is 2601 million tons. The inclusion of cereals in our diet about 11,000 years ago, played an important role in human evolution and social interaction. Wheat (Triticum aestivum) is the most widely cultivated crop on Earth, contributing to about one fifth of the total calories consumed by humans.2 Today, wheat cultivation is widespread and, together with maize and rice, has become one of the most important crops in the globe. Approximately 600 million tons are harvested annually with cultivation extending over a vast geographical area.1,3 Consequently, wheat yields and production affect the global economy, and failed harvests can lead to social unrest. Wheat has also an important nutrition value because it is a rich source of vitamins, proteins and carbohydrates in its natural form.1 However, the prevalence of wheat-related disorders has increased in the last decades.4–6 These involve a wide spectrum of conditions, triggered by the ingestion of gluten-containing cereals such as celiac disease (CeD), gluten ataxia, some cases of dermatitis herpetiformis, IgE-mediated reactions such as wheat allergy, or the poorly characterized gastrointestinal symptomatic condition termed non-celiac gluten/wheat sensitivity (NCWS).4,7,8
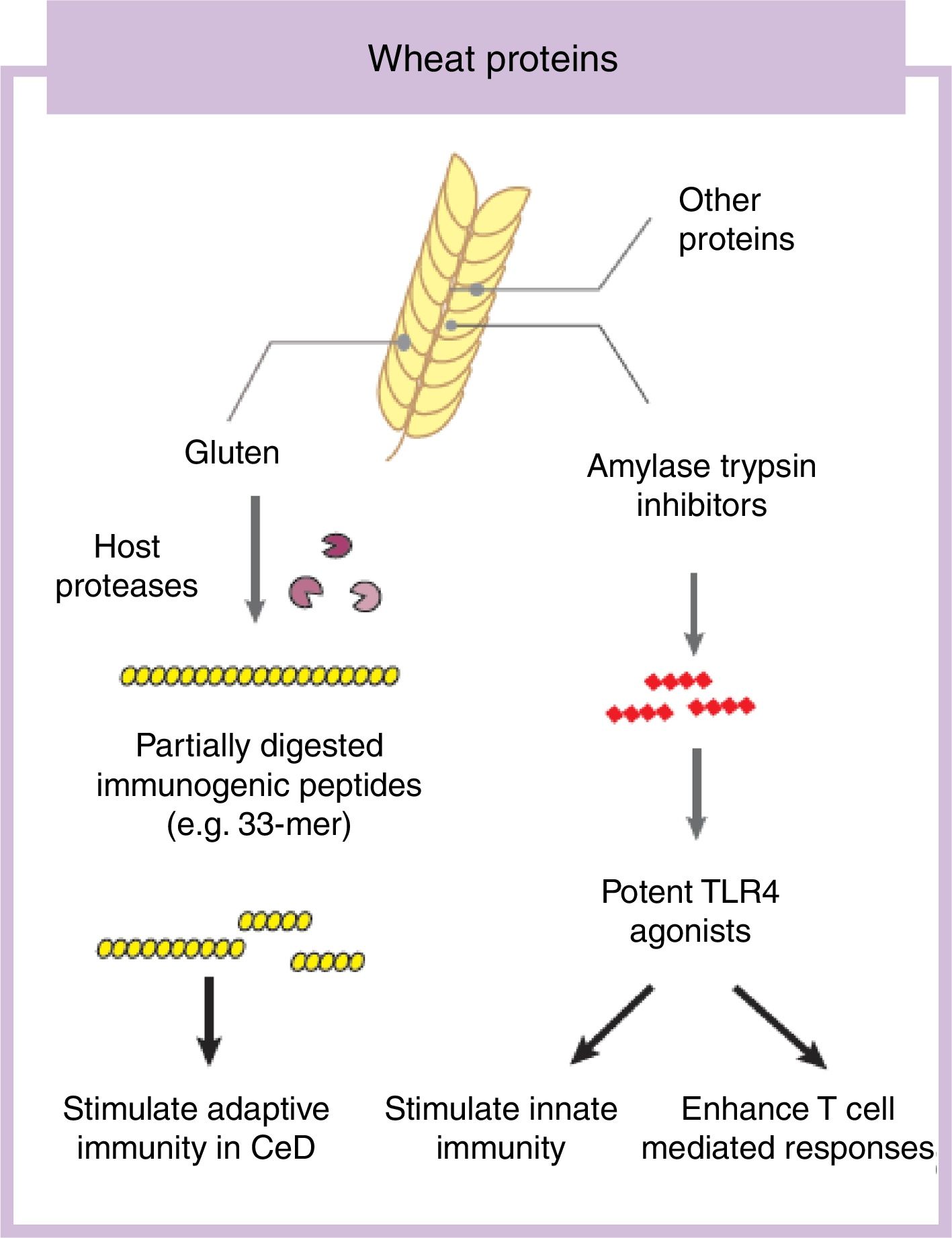
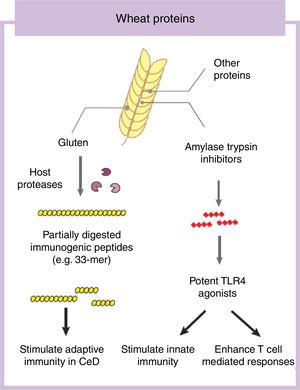
Some components in cereals, and specifically in wheat, have the capacity to induce adverse reactions in people.9 Recently the role of fermentable non-protein components such as oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAP) have gathered considerable attention despite a clear understanding of mechanisms of action.10,11 In this review we will focus on the protein fraction in wheat, which may incite food sensitivity particularly in individuals with a certain genetic predisposition (Fig. 1). We will concentrate on the well characterized entity CeD, whose main environmental trigger is gluten in wheat, rye and barley as well as the controversial entity called NCWS, in which the exact wheat trigger component is not clear.12,13 For the specific case of wheat, the protein fraction can be classified into albumins, globulins, gliadins, and glutenins, according to their structural properties and solubility.3 As we discuss below, gluten is the global term we give to a type of proteins (prolamins) contained in the storage fraction of wheat, rye and barley grain.14 Gluten includes gliadins and glutenins in wheat and similar proteins, hordeins and secalins, are present in rye and barley respectively.15 It has been speculated that other non-gluten proteins in wheat could lead to adverse reactions.13 Among these, amylase trypsin inhibitors (ATI) can stimulate innate immune responses that could play a role in wheat sensitive disorders16 (Fig. 1).
The prevalence of wheat-related disorders has increased alarmingly in the last decades.4,6 Although it is well established that a genetic predisposition is needed for defined conditions such as CeD to develop,17,18 the increase in prevalence and the change in clinical spectrum has been too quick to be explained only by genetic drift, pointing toward a change in environmental exposures as risk modifiers.19 Several factors have been proposed, and among these the role of intestinal microbes has emerged.19 In the next sections, we will discuss the role of specific protein fractions from wheat in the gastrointestinal conditions included in the wheat related disorder spectrum. We will also discuss the interactions of these agents with intestinal microbes with relevance in disease.
Proteins of relevance in wheat related disordersGluten fractionGluten is defined as the remaining mass when wheat dough is removed of starch granules and water-soluble constituents; which include the water insoluble protein fraction, lipids (up to 10%) and some water insoluble starches.14 This mass contains 75–85% proteins that play a key role in determining the unique baking quality of wheat by conferring water absorption capacity, viscosity and elasticity on the dough.14 These characteristics have raised gluten as an important ingredient in food industry.1 Based on the solubility, gluten proteins are divided into gliadin (soluble in ethanol 60%) and glutenins (non-soluble in ethanol).14 The gliadin fraction consists of highly homologue monomeric proteins that are divided into α/β-, γ- and ω-gliadins according to electrophoretic migration.14,15 Glutenins are high-molecular weight aggregated proteins responsible for the dough elasticity.14 However, from a medical perspective the term “gluten” is not limited to wheat proteins. Similar proteins presented in phylogenetically close related cereals such as rye and barley (hordeins and secalins) are also included in the term gluten.12 The potential role of similar proteins (avenins) in oats as triggers of inflammation in CeD has been addressed in previous studies20–22 and in a metaanalysis.23
Gluten is the main and necessary environmental trigger of CeD, the most important and best characterized condition within the umbrella of wheat-related disorders.12 Gluten proteins are rich in proline and glutamine amino acid residues conferring an unusual resistance of these proteins to mammalian proteases.24 As a consequence, large and highly immunogenic gluten peptides appear in the gastrointestinal lumen. These peptides can induce an adaptive25 and/or, although more controversial, innate immune response in the small intestine of humans.26,27 For the adaptive immune response, partially digested gluten peptides such as 33-mer, with 3 well-described immunogenic epitopes (PYPQPQLPY, PQPQLYPQ, PFPPQPQLPY)24 translocate the mucosal barrier where they are deamidated by human transglutaminase 2, the CeD-associated autoantigen.28 This process converts glutamine residues to glutamate and increases peptide binding affinity to human leukocyte antigen (HLA)-DQ2 or DQ8 heterodimers in antigen-presenting cells, initiating the characteristic CD4 T-cell response in CeD.25 Different authors have shown that innate immune activation is also needed to develop enteropathy in CeD (gold standard for diagnosis) which includes an increase of intraepithelial lymphocytes, crypt hyperplasia and villus shortening in the duodenum of the patients.12 Several entities have been proposed as activators of the innate immune response including specific gluten peptides such as 19-mer.26,27 The effects of 19-mer in inducing an innate immune response seem to be dependent on myeloid differentiation primary response 88 (MyD88) and type I interferons but more studies are needed to understand the exact implications in CeD.29
Gluten is frequently associated to other wheat-related disorders such as ataxia and dermatitis herpetiformis, but the specific mechanisms are not as well as characterized as in CeD.12 The role of gluten in other wheat related disorders such as NCWS or wheat allergy is more controversial.13 Although patients with NCWS frequently recognize gluten as the main trigger of their symptoms, other components in wheat could act as main activators, which will be discussed in the next section.
Non-gluten fraction: wheat amylase trypsin inhibitorsThe non-gluten protein fraction in wheat is constituted by albumins and globulins that contain up to 14 families of proteins, such as the ATI, β-amylases, peroxidases, lipid transfer proteins, serpins, and others.30 Among these, wheat ATI are largely resistant to intestinal proteolysis and with the capacity to trigger innate inflammation by engaging the toll-like receptor 4 complex on myeloid cell16 (Fig. 1). ATI are a family of at least 11 structurally similar proteins highly presented in modern wheat, and in less proportion in rye and barley, which serve as protective proteins by inhibiting enzymes (amylase and trypsin-like activities).30 The activation of innate immunity by ATI leads to gut dysfunction, low grade inflammation and an increase in intestinal permeability which can exacerbate pre-existing colonic and small intestinal inflammation as it has been shown in colitis and gluten-sensitivity mouse models.31,32 This response is independent of genetic status and despite the absence of mucosal damage, suggesting that ATI could participate in some forms of wheat-related disorders such as NCWS.31 In contrast, other ATI-like molecules present in other staples different than wheat are inactive.32 When combined with gluten in a permissive genetic background, wheat ATI exacerbates gluten-induced immunopathology, suggesting an adjuvant role to gluten in wheat-related disorders such as CeD.31 The inflammatory signal led by ATI can be also propagated to the mesenteric lymph nodes affecting antigen priming and playing a role in extraintestinal manifestations of wheat sensitivity.32 For instance, it has been shown that wheat ATI could be important nutritional activators and adjuvants of allergy.33,34
Other potential wheat proteins of relevance are wheat germ agglutinins (WGA). WGA are enriched in the germ of wheat grains with contents from 100 to 500mg/kg, resulting in typical concentrations of approximately 4mg/kg in white flour and approximately 30mg/kg in whole grain flour.35 Similar to ATIs, WGA are resistant to proteolysis by mammalian proteases and has been shown to induce the release of proinflammatory cytokines such as TNF-alpha, IL-1B, IFN-gamma and IL-12.36 However, no stimulatory activity has yet been demonstrated in animal models or clinical practice.13
The role of intestinal microbiota in wheat-related disordersThe human intestine harbors a complex community of over 100 trillion microbial cells which influence human physiology, metabolism, nutrition and immune function. Disruption of this interaction has been linked with inflammatory conditions and metabolic disorders.37,38 Of relevance for this review, several studies have shown differences in the intestinal microbiota composition of patients suffering from CeD compared to healthy controls.39,40 Although altered microbiota in these patients could be either consequence of their dietary habits (e.g. gluten or wheat-free diet) or the disease process (inflammation, malabsorption), several studies have shown that intestinal microbiota could play a primary or inductive role,19 as we explain below. We will discuss in the next subsections the most important diet–microbe interactions of relevance for wheat related disorders.
Microbiota alterations in celiac diseaseThe role of enteric microbes in inducing specific food sensitivities and autoimmune disorders has been proposed.19 Because CeD is the best characterized wheat-related disorder and several studies have investigated intestinal microbiota structure in these patients, we will focus on this entity in this section. Considerably less information is available regarding the role of the intestinal microbiota in other wheat-related disorders such as NCWS or wheat allergy. The difficulty in accurately diagnosing NCWS, and in identifying the exact trigger in wheat responsible for these conditions adds a layer of complexity.13
Although not all the studies are conclusive, it seems that patients with active CeD present different microbiota profiles when compared to healthy or non-CeD volunteers.39 However, the study of the intestinal microbiota in CeD is not easy for several reasons. First, the most relevant studies are those performed in duodenal biopsies, the most affected area in CeD. The alterations in the fecal microbiota and in fecal bacterial metabolism described in CeD patients compared with healthy volunteers are important as they provide an overall snapshot of the whole microbiota in that population.41–43 However, those analyses may miss changes in microbiota that are associated with local proximal duodenal inflammation or find others that are not causally related with the celiac process. On the other hand, the malabsorption syndrome associated with CeD12 could cause a different fecal environment, leading to secondary changes in fecal microbiota that are not necessarily a direct reflection of the communities in duodenal biopsy. Second, microbiome analysis in the small intestine is rarely performed in true healthy volunteers as it requires an invasive gastroduodenal endoscopy. As a consequence, patients with functional symptoms who attend the clinic and in whom CeD is ruled out, are often recruited as healthy “disease” controls. Last, but equally important, the analysis of intestinal microbiome in the small intestine is not as straightforward as in other parts of the gastrointestinal tract. Many studies in CeD have been performed using first generation techniques for microbiota determination such as quantitative PCR, denaturing gradient gel electrophoresis (DGGE) or temperature gradient gel electrophoresis (TGGE), culturing and clone collection.44–48 These studies have provided very valuable information but now require to be validated by using high throughput sequencing techniques and metagenomics that allow studying the microbiota at a deeper level.49,50 The new sequencing techniques of microbiota have revolutionized the comprehension of diet–microbiota and host–microbiota interaction in different disorders.38 However, the use of these techniques in samples with a low yield microbiota such as duodenal biopsies is difficult and requires specific expertise.51 Duodenal biopsies contain more host than bacterial DNA, and contaminations coming from reagents or sample scooping can compromise the results. In addition, the sequence deepness is lower than when using fecal or colonic samples.49 This needs to be taken into consideration when attempting to analyze and interpret mucosal (duodenal)-associated microbiota in CeD.
Despite this, several studies suggest an important role of microbes in CeD.39 Environmental modifying factors of the intestinal microbiota such as antibiotic intake, C-section delivery in pregnancy or proton pump inhibitor consumption have been associated to CeD.52–54 Although the large cohort studies have not confirmed these independent observations,55,56 it is important to say they did not account always for differences into classes of antimicrobials, duration and reasons for their use, and between elected or emergency C-section procedures. In addition, different groups have described changes in the intestinal microbiota of CeD patients compared with controls.39,40,50 The duodenal microbiota composition of CeD patients have been also associated with the clinical manifestation of the disease.57 More challenging is to define the characteristics of a healthy or a pathogenic duodenal microbiota associated to CeD. Similar to other chronic inflammatory conditions, CeD patients have been reported to have an increase in the abundance of Proteobacteria with proliferation of opportunistic pathogens such as Neisseria, E. coli or Pseudomonas, as well as higher bacterial virulent genes.42,50,58–60 An increased presence of pathogenic bacteria is also observed in the gut of infants at risk of developing CeD.61 The genotype of infants at risk of developing CeD, carrying the HLA-DQ2 haplotypes, influences the early gut microbiota composition.62 Thus, alterations in the early trajectory of gut microbiota in infants at CeD risk could influence the immune maturation process and predispose to CeD.63 The metabolic capacity of the intestinal microbiota has also been studied in the context of CeD, with active CeD patients presenting differences in specific bacterial metabolites and an increase in bacterial proteases.41,60,64,65 We have recently shown that duodenal biopsies of CeD patients have a high proteolytic activity that correlates with the proliferation of specific opportunistic pathogens such as Pseudomonas in the small intestine with the capacity to induce gluten-independent inflammation in the host.60 Of mention, after a strict gluten-free diet, only those CeD patients responding to the treatment present a similar microbiota as non-CeD controls, highlighting the importance of the intestinal microbiota in the disease course and responses to treatment.57 Although these data are associative, animal studies suggest that microbes could participate in the development of gluten sensitivity by different mechanisms, as we have previously explained in a recent review.19 Understanding these mechanisms will open a new avenue for treatment or diagnosis of the disease.
Does a wheat-free diet impact the microbiota and how?Historically, a wheat- or gluten-free diet was recommended only for those patients diagnosed with CeD or IgE-mediated wheat allergy. Today, the wheat-free diet is adopted in the absence of a CeD or wheat allergy diagnosis and sometimes in otherwise healthy people.66 As such, this diet is advocated in weight loss and energy programs, despite a consistent lack of evidence to support their use and effectiveness or even, its long-term consequences. The increasing demand for gluten-free products reflects a popular misconception among consumers that gluten avoidance is part of a healthy lifestyle choice. The vilification of wheat in non-peered reviewed literature has created an empire founded on the premise that gluten is a poison, reinforcing powerful myths and promises of simple dietary solutions to numerous health problems.66,67 Since microbiota stability and richness has been associated with health,38 does wheat, barley and rye restriction affect the intestinal microbiota of healthy adults?
In a recent study involving healthy adults, Hansen et al.68 found that a low-gluten diet induces moderate changes in the intestinal microbiome, reduces fasting and postprandial hydrogen exhalation, and leads to reduction in self-reported bloating; suggesting that low-gluten diet in healthy adults may be driven by qualitative changes in dietary fibers.68 In addition, functional activity of the intestinal microbiota is modified by gluten intake in the diet of healthy volunteers.69 However, the gluten-free diet also leads to reductions in beneficial gut bacteria populations, modification of their normal metabolic capacity and their ability to stimulate host's immunity.70,71 It is important to mention that exclusion of certain components from the diet in healthy people could lead to loss of groups of bacteria, or bacterial functions that efficiently degrade and detoxify the dietary component, as they will adapt to a different dietary source.19 Thus, traditional dietary advice rather than excessively restrictive diets may be better suited for maintaining microbiota fitness (or richness),72 and there is no strong evidence that a gluten or wheat-free diet in the absence of a diagnosis of CeD is beneficial.
When a diagnosis of CeD is made, complete removal of gluten from the diet is necessary which allows for mucosal healing and significant clinical improvement.73 Although differences in the upper small intestinal microbiota between treated and untreated CeD in adults have been described,44 these differences could be the consequence of mucosal healing, in addition to the withdrawal of gluten from the diet. However, about 30% of patients can remain symptomatic, which may indicate deficiencies in the gluten-free diet or may prompt for additional diagnosis. Recently, the role of persistent microbial changes in the upper gastrointestinal tract of CeD patients who were non-responders to the gluten-free diet was described.57 The mechanisms through which dysbiosis could maintain symptoms in CeD patients after the gluten-free diet are not clearly understood.
There are non-gluten wheat proteins, such as ATI, that are removed from the diet during a gluten-free diet. Animal studies have shown that gluten intake has the capacity to shape the intestinal microbiota and ATI intake has been associated to changes in specific bacterial groups.31 This has relevance to wheat-related disorders which may be caused or potentiated by reactions to non-gluten protein fractions. This was shown in a recent study where a wheat-free diet changed microbiota composition in mice, but when ATI were added to the wheat free diet they cause gut dysfunction.31 Moreover, when ATI were combined with gluten in a mouse model expressing celiac genes HLA-DQ8, they exacerbated inflammation and lead to changes in the intestinal microbiota relevant for CeD.31 However, more studies are needed to decipher the importance of wheat-free diet and the individual and combined role of wheat proteins in shaping composition and metabolic capacity of the human intestinal microbiota.
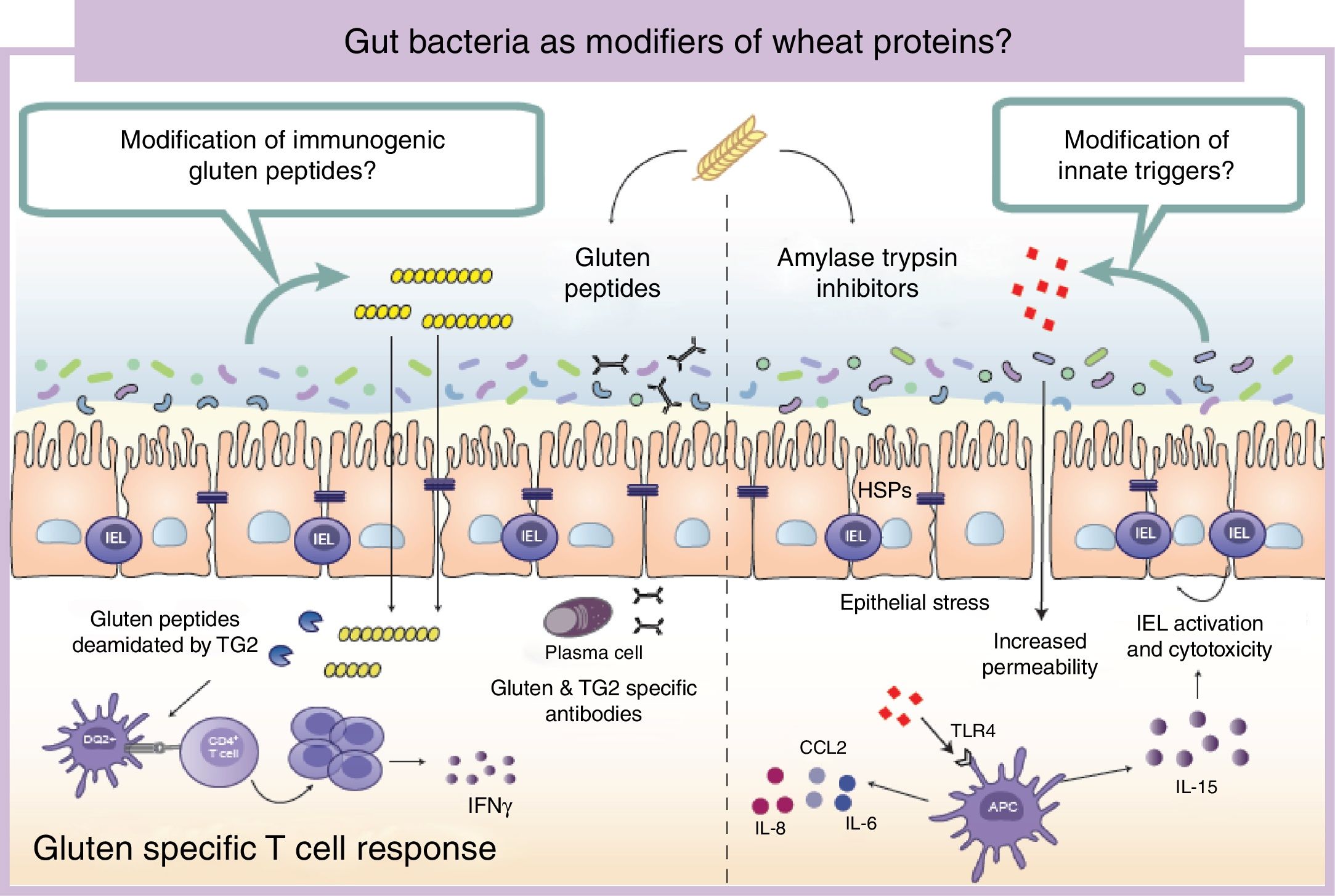
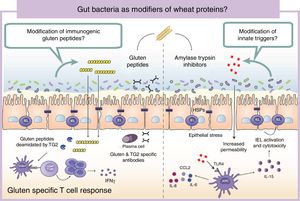
Wheat protein metabolism by the microbiotaImmunogenicity of wheat proteins such as gluten and ATI are defined by their resistance to mammalian digestive proteases.24 As a consequence of this partial digestion, large and immunogenic peptides capable of inducing adaptive and/or innate immune responses are generated in the human gastrointestinal tract. However, the role of intestinal microbiota in gluten and ATI degradation has frequently been underestimated.19 The functional diversity of the human gut microbiota implies a vast catalog of metabolic pathways74 that participates in the digestion of dietary components, even those that are difficult to digest by human digestive enzymes.19 Dietary components not used by the host become substrates for intestinal microbes that ultimately will dictate their immunogenicity.75 The human gastrointestinal tract harbors microbes with the ability to digest gluten and ATI31,76–80 (Fig. 2). It is important to mention that some of these microbes are present in the oral cavity and in the proximal small intestine suggesting that an efficient degradation of the peptides at those sites could reduce immunogenicity in the second portion of the small intestine, the most affected area in CeD.76,79 We have shown that some intestinal microbes can reduce immunogenicity of gluten peptides in mouse models.75 We have also found that intestinal microbes can efficiently degrade ATI and in that way reduce their capacity to induce gut dysfunction and innate immune activation in mice.31 It can therefore be hypothesized that healthy people with genetic predisposition to certain wheat-related disorders could be protected if they harbor a metabolically efficient intestinal microbiota with optimal dietary protein modification. Of note, differences in the metabolic capacity of the intestinal microbiota toward wheat protein substrates have been reported between patients with CeD, relatives of CeD and healthy subjects.64 Specific Lactobacillus strains, a group depleted in CeD patients and frequently used as probiotic, have the capacity to efficiently degrade both gluten and ATI proteins, reducing their immunogenic characteristics invivo.31,75,80 These results have opened new directions for therapeutic development using small intestinal bacteria or specific proteases with optimal gluten degrading capacity.72,81
Gut microbiota as modifiers of wheat proteins. The human intestinal microbiota contains bacteria with the ability to degrade and change dietary antigens such as gluten or amylase trypsin inhibitors modifying their immunogenic potential and affecting the susceptibility of the host to develop wheat related disorders. APC: Antigen presenting cell; CCL: Chemokine (C-C motif) ligand; HSP: Heat shock proteins; IEL: Intraepithelial lymphocyte IFN: Interferon; IL: Interleukin; TLR4: Toll-like receptor 4.
It is of outmost importance to clarify that the terms “immunogenic protein metabolism” or “immunogenic protein degradation” does not always indicate detoxification, and merely points at a metabolic capacity of that bacteria toward the substrate.19 We found that the intestinal microbiota has a dual effect in gluten metabolism, increasing or reducing gluten immunogenicity. Some microbes have the ability to degrade non-digested gluten peptides producing shorter peptides with retained or higher immunogenicity that translocate efficiently the mucosal barrier.75 In addition, our group has recently shown that some bacterial proteases that degrade gluten peptides can directly induce an innate immune response characterized by an increase of intraepithelial lymphocytes and up-regulation of genes relevant for CeD such as interferon gamma or fasl in animal models.60 When mice express specific celiac risk genes (e.g. HLA-DQ8) that recognize gluten peptides, the activation of the innate immunity by specific bacterial proteases lead to more severe immune-pathology characterized by a reduction in the villus-to-crypt ratio.60 It is important to mention that an increase in proteolytic activities toward gluten peptides, and a reduction in specific protease inhibitors, has been previously described in CeD patients.64,82–84 Of interest, serine protease inhibitor produced by a probiotic strain is able to reduce gluten-induced immunopathology in a mouse model.85 Thus intestinal microbiota is an important factor that can determine wheat protein immunogenicity, and through this mechanism could influence risk to develop wheat related disorders.
ConclusionsThe interest in wheat-free foods has significantly increased in the past ten years, and only in North America, a staggering 11 million people are following this diet based on the assumption that gluten is deleterious for health, rather than on a justified medical diagnosis, such as celiac disease. This is why mechanistic studies to determine whether and how different wheat components induce symptoms or immune pathways in individuals without celiac disease are needed. On the other hand, the study of microbial factors as additional determinants of risk for defined conditions such as CeD, or other wheat-induced reactions, could shade some light into the increasing prevalence of these conditions as well as promote research into new therapies aimed at improving the metabolic capacity of the intestinal microbiota toward immunogenic wheat proteins.
Conflict of interestsThe authors declare no conflict of interests.
Grant supportThe work was supported by a CIHR grant to EFV MOP# 142773. EFV holds a Canada Research Chair. AC holds a Farncombe Family fellowship and Biocodex microbiota Foundation Award.