We are currently living in an unprecedented era of hepatitis C treatment with the availability of highly effective drugs yielding minimal side effects. The problem we currently face is the retreatment of patients refractory to these drugs. Although several factors can influence treatment failure, this review focuses on antiviral resistance. Resistance-associated substitutions may be identified at baseline or be treatment-emergent. The latter seem to be more clinically relevant and must be studied in the event of treatment failure (no virological response). In this article, we present the latest data from clinical trials and studies in real-life clinical practice. Finally, based on this current evidence, we propose some recommendations for the management and retreatment of these patients.
En el momento histórico en el que nos encontramos en el ámbito del tratamiento de la hepatitis C, en que disponemos de fármacos excepcionalmente eficaces y con muy escasos efectos secundarios, se nos plantea el problema del retratamiento en los pacientes que fracasan al mismo. Los factores que influyen en el fracaso son muy diversos, si bien en esta revisión nos vamos a centrar en las resistencias antivirales. Las sustituciones asociadas a resistencias pueden ser tanto basales como inducidas por el tratamiento; estas últimas parece que son las más importantes clínicamente y las que deben ser estudiadas ante la falta de respuesta virológica. En este artículo ofrecemos los últimos datos de ensayos clínicos y estudios en práctica clínica real, y en base a la evidencia actual se ofrecen unas recomendaciones de manejo y retratamiento de estos pacientes.
Direct-acting antivirals (DAAs) are currently the treatment of choice for hepatitis C virus (HCV) infection.1 These drugs achieve a sustained virologic response (SVR) rate of over 90% in virtually all population groups, and only slightly lower in cirrhotic patients with genotype 3 infection in whom prior treatment has failed. DAAs also present an excellent safety profile, especially in regimens that do not include ribavirin (RBV).
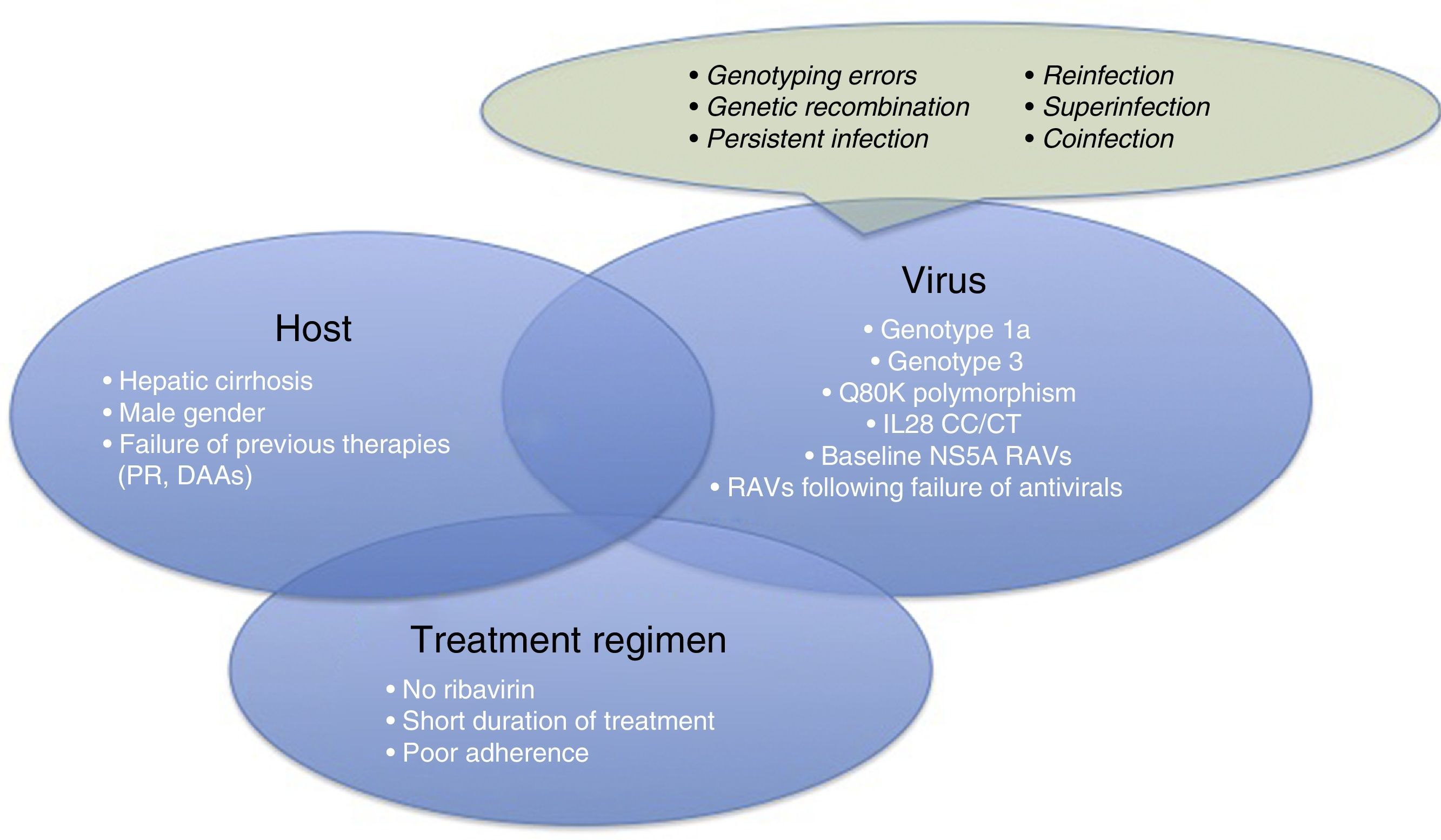
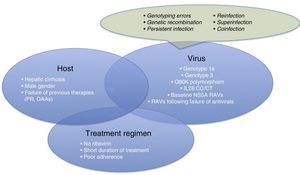
Therefore, it is time for us to turn our attention to therapeutic failures, which can be due to various host-, virus-, or antiviral-related factors (Fig. 1). One of the most interesting causes of treatment failure is the development of resistance-associated variants (RAVs). Accurate RAV testing is essential. Understanding of RAVs has improved remarkably, and we now know that not all are clinically significant, and not all need to be analysed.
This review aims to provide an in-depth analysis of RAVs, what they are, their clinical importance, when they appear, when to test for RAVs or not, and by what method. This analysis of RAVs will enable clinicians to select the best re-treatment regimen for their patients.
Hepatitis C virus. Virological concepts and variabilityHCV is a single-chain RNA virus belonging to the Hepacivirus genus of the flaviviridae family of viruses discovered in 1989.2 HCV circulates in the blood in the form of lipo-viro-particles. Once it enters the hepatocyte, the RNA is released and begins to synthesise the polyprotein that will be processed by viral (NS2-3 and NS3-4A) and host proteases. New copies of RNA are formed in the viral replication complex, which is composed of non-structural proteins (NS3, NS4A, NS4B, NS5A and NS5B). As a result, intracellular membranes are reorganised to generate an ideal viral replication micro environment called the membranous web, composed of double-membrane vesicles. Finally, assembly takes place in the endoplasmic reticulum.
HCV is an exceptionally variable virus,3 thanks to its high estimated turnover rate of 2–5h, and its enormous daily virion output (1012 virions) per patient. Added to this is its high rate of mutation (10−4 to ×10−5 per nucleotide and per replicative cycle) due to the low error-repair capacity of RNA-dependent RNA polymerase.4,5
This gives rise to the large number of HCV variants, most of which are directly eliminated by the immune system or are unable to replicate due to loss of coding proteins. Others, however, are able to survive and perpetuate the infection.
The variants that survive give rise to the 7 different genotypes, differentiated by a 30%–35% variation in nucleotide sequence, and 67 subtypes, whose genomic sequence varies by between 20% and 25%. In addition, a dominant species (wild type) and different quasispecies, with a sequence variability of up to 10%, can be found in the same patient.
Causes of therapeutic failureFailure of antiviral treatments may appear at different times:
- •
During antiviral treatment, it is called breakthrough.
- •
Once the treatment is completed, it is called viral relapse. This is most common in patients receiving DAAs.
- •
Null response to antiviral treatment, primary absence of response to treatment.
As explained above, modern antivirals are extremely effective. However, 1%–7% of patients are not able to be cured.6 This is due to many different reasons.
Before focussing on resistance-related HCV treatment failures, we will briefly explain the other reasons for therapeutic failure. These are summarised in Fig. 1.
- •
Patient-related causes: male gender, compensated cirrhosis, failure of previous treatments (based on interferons [IFNs] or DAAs)
- •
Virus-related causes: genotype 1a, 3, baseline NS5A RAVs, RAVs developed after treatment with DAAs, Q80K (only in patients receiving treatment with simeprevir [SMV]), unfavourable IL28b polymorphism.
There are other causes of virological failure aside from RAVs. These include genotyping errors, genetic recombination phenomena (rare cross-linking of RNA intermediates leading to recombination of 2 viral strains, for example, 1 hybrid virus with genotype 1 and 2), persistent infections, reinfections, superinfection (combination of reinfection and persistent infection evidenced in the phylogenetic analysis of the virus), or coinfections.
- •
Treatment regimen-related causes: lack of adherence to treatment, short treatments, no RBVs.
For virological failure to occur, 2 or more of the aforementioned factors must be present in the same patient,6 although the presence of each can have a discrete effect on achieving SVR.
Consequences of viral variabilityThe concept of RAVs, which we will explore in greater depth further on, arises from the ability of the HCV to produce variants, which are, in short, changes in the nucleotide sequence responsible for the synthesis of proteins targeted by different DAAs (NS3/4A, NS5A, NS5B).
The ability to generate variants, and in particular to produce RAVs, is an important factor in patients who do not respond to DAA therapy. This is because a DAA that is not sufficiently potent and only partially inhibits viral replication can facilitate selection of variants and the appearance of RAVs that may reduce viral susceptibility to future antiviral treatments.
DAAs act on key points in the HCV cell cycle, and derive their name from the target on which they act. The DAAs which are currently available are shown in Table 1.
- •
Non-nucleoside NS5B polymerase inhibitors: dasabuvir (DSV).
- •
Nucleoside NS5B polymerase inhibitors: sofosbuvir (SOF).
- •
NS3/4A protease inhibitors: SMV, ritonavir-boosted paritaprevir (PTVr) and grazoprevir (GZV).
- •
NS5A inhibitors: ledipasvir (LDV), elbasvir (EBV), daclatasvir (DCV), velpatasvir (VPA), ombitasvir (OBV).
Currently available direct antiviral agents (DAAs).
| Therapeutic target | Drugs |
|---|---|
| Non-nucleoside NS5B inhibitors | Dasabuvir (DSV) |
| Nucleoside NS5B inhibitors | Sofosbuvir (SOF) |
| NS3/-4A protease inhibitors | Simeprevir (SMV), ritonavir-boosted paritaprevir (PTVr), grazoprevir (GZV) |
| NS5A inhibitors | Ledipasvir (LDV), elbasvir (EBV), daclatasvir (DCV), velpatasvir (VPA), ombitasvir (OBV) |
DAAs act on several viral targets in an attempt to completely suppress viral replication, and thus, achieve cure.
It is logical to assume that combining different DAAs with different viral targets is the best way to prevent the emergence of multidrug-resistant viruses.
Therefore, standard treatment regimens usually consist of a polymerase inhibitor accompanied by 1, 2 or 3 drugs that act on different targets.
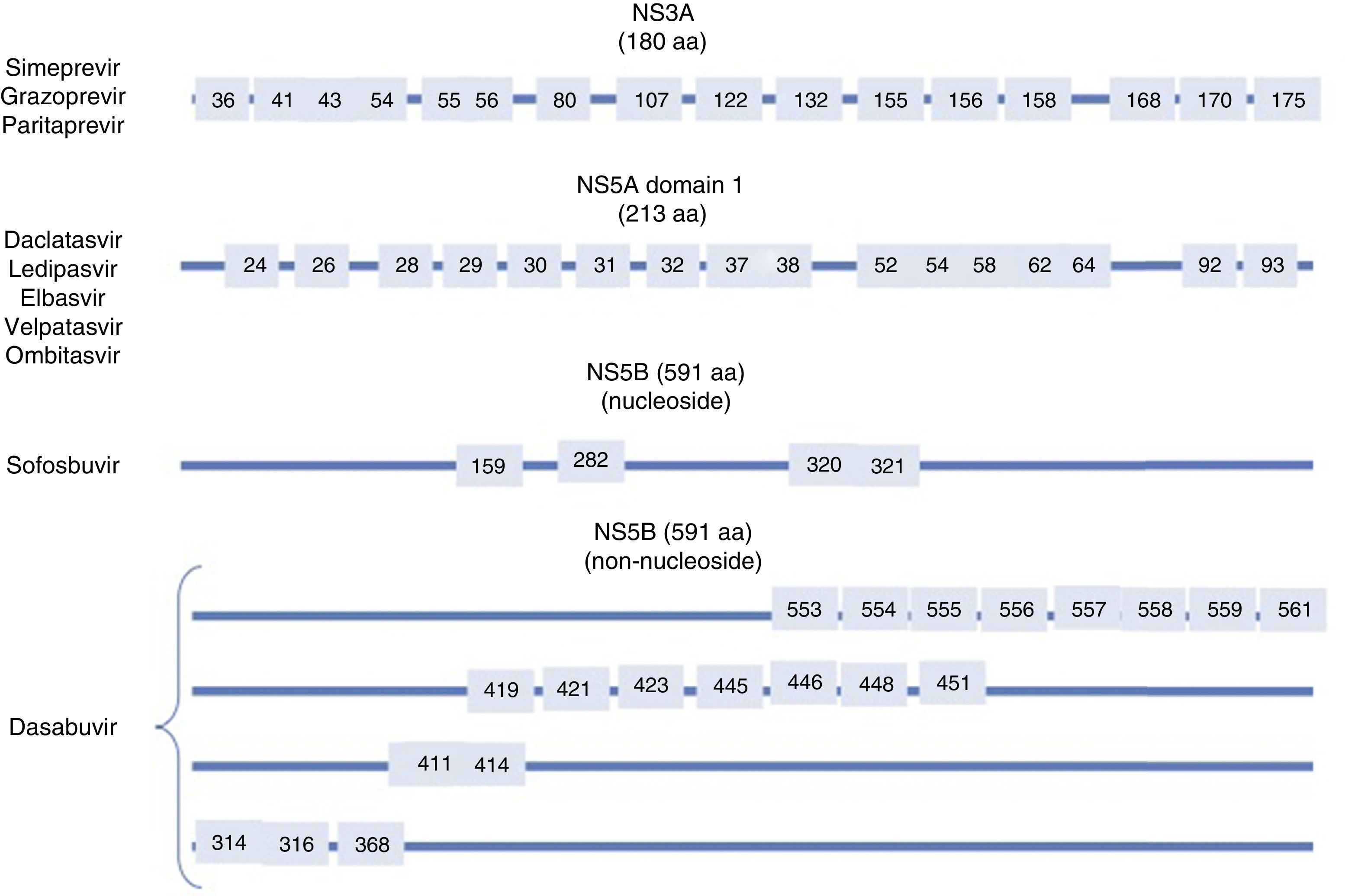
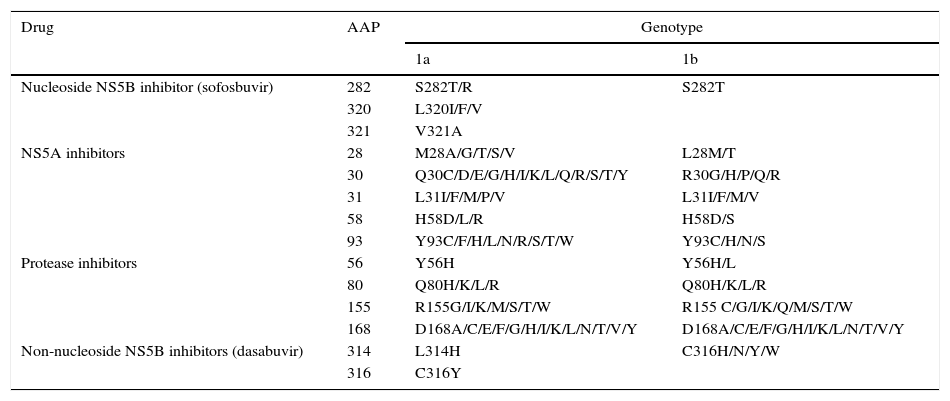
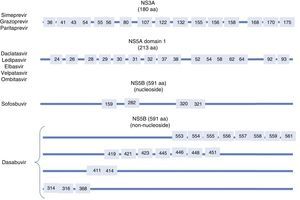
Resistance-associated variantsAs mentioned above, RAVs are a result of HCV variability, which generates mutations in the nucleotide sequence responsible for the synthesis of the proteins targeted by different DAAs (NS3/4A, NS5A, NS5B) (Fig. 2). HCV is known to have a greater capacity to generate RAVs than other viruses, such as HIV or HBV.7 The very presence of RAVs, however, does not explain resistance to a treatment. Resistance to, and ultimately failure of, treatment depends on both virological and non-virological factors. The following are factors implicated in the importance of RAV-related resistance: 1) the quantitative importance of the RAV, 2) RAV type: baseline/DAA failure-related, 3) the potency of the antiviral regimen, 4) the DAA's genetic barrier to resistance: number of mutations necessary for HCV to become DAA-resistant. SOF is the antiviral with the greatest genetic barrier, and 5) Viral fitness, i.e., its replicative capacity. The most common RAVs are shown in Table 2.
Most important resistance-associated variants (RAVs).
| Drug | AAP | Genotype | |
|---|---|---|---|
| 1a | 1b | ||
| Nucleoside NS5B inhibitor (sofosbuvir) | 282 | S282T/R | S282T |
| 320 | L320I/F/V | ||
| 321 | V321A | ||
| NS5A inhibitors | 28 | M28A/G/T/S/V | L28M/T |
| 30 | Q30C/D/E/G/H/I/K/L/Q/R/S/T/Y | R30G/H/P/Q/R | |
| 31 | L31I/F/M/P/V | L31I/F/M/V | |
| 58 | H58D/L/R | H58D/S | |
| 93 | Y93C/F/H/L/N/R/S/T/W | Y93C/H/N/S | |
| Protease inhibitors | 56 | Y56H | Y56H/L |
| 80 | Q80H/K/L/R | Q80H/K/L/R | |
| 155 | R155G/I/K/M/S/T/W | R155 C/G/I/K/Q/M/S/T/W | |
| 168 | D168A/C/E/F/G/H/I/K/L/N/T/V/Y | D168A/C/E/F/G/H/I/K/L/N/T/V/Y | |
| Non-nucleoside NS5B inhibitors (dasabuvir) | 314 | L314H | C316H/N/Y/W |
| 316 | C316Y | ||
AAP: amino acid position.
- -
Baseline RAVs: these are RAVs detected in previously untreated or treatment-naïve patients. Their frequency depends on the HCV genotype and subtype.3 Baseline RAVs do not appear to influence SVR.
- ∘
Baseline NS3 RAVs are found in 3% of patients. It should be noted that the presence of Q80K polymorphism, found in more than half of all patients infected by genotype 1a,8 is a predictor of lower SVR rates in patients treated with pegylated interferon (IFN), RBV and SMV. However, after the development of DAAs, the presence of this polymorphism at baseline has less effect on combinations of SOF+SMV, and baseline determination is only recommended in patients with cirrhotic genotype 1a scheduled for treatment with SOF+SMV.9
- ∘
Baseline NS5A RAVs have no clinical importance when deciding on treatment, unless they appear after a previous treatment. In some studies in patients treated with SOF+LDV, baseline LDV RAVs reduced SVR rates in treatment-experienced patients with no exposure to NS5A inhibitors.10 Attention should be drawn to EBV RAVs; population genomics performed in the context of phase II/III trials for EBV/GZV approval have shown that in order to achieve high SVR rates with 12-week regimens in cirrhotic HCV genotype 1a patients with previous null response, it is necessary to rule out the presence of EBV-resistance baseline RAVs in NS5A (variants at positions 28, 30, 31, 93), as these reduce susceptibility to this antiviral by five-fold or more.11
- ∘
Finally, several baseline NS5B RAVs have been described for both DSV and SOF, but these are extremely infrequent and seem to have little impact on viral response. Some SOF RAVs, such as C316N/H/F and the S282T variants, have been found in a small percentage of patients, but their effect on the antiviral response still needs to be clarified.12
- ∘
A study published recently, in which 312 European patients participated, investigated the baseline RAVs of the NS3, NS5A and NS5B regions that were potentially relevant to treatment with telaprevir, SMV, asunaprevir, DCV, LDV, OBV and DSV. No SOF-resistant RAVs were observed at baseline. RAVs to NS3 were detected in 20.5% of cases, to NS5A in 11.5% (more frequent in genotype 1b than in 1a), and to NS5B in 21.5% (also more frequent in genotype 1b). The authors concluded that baseline RAV testing should be performed prior to starting treatment.13 Although, in line with the previous study, some authors conclude that antiviral therapy should be guided by baseline RAVs, especially in cirrhotic patients or those scheduled for shorter regimens,14 there is currently no evidence to support this, and no clinical guidelines recommend such tests. Studies in patients treated with SOF+LDV for least 6 weeks or retreated for 12 weeks after treatment with shorter regimens achieved the same SVR rate with or without RAVs.15,16 In another similar study, baseline NS5A RAVs were detected in 94 cases (of a population of 511, 18% of which were cirrhotic patients) treated with SOF and LDV. SVR rates were similar in patients with and without baseline RAVs (91 vs 98%).17 Testing for Q80K polymorphism in the NS3 region should only be recommended prior to administration of SMV+SOF in cirrhotic HCV genotype 1a patients,18 and baseline RAVs should only be determined in HCV genotype 1a patients (in particular in positions M28, Q30, L31 and Y93) scheduled to receive EBV/GZV treatment. If the variant is detected, RBV should be added and treatment extended to 16 weeks.19
- ∘
- -
RAVs that appear in patients who have failed previous antiviral treatment:
- ∘
NS3 RAVs: these are found in <7% of treatment-experienced patients.20 They are usually transient, and disappear shortly after suspension of the drug due to their low replicative capacity (viral fitness).20 The RAVs most frequently associated with therapeutic failure emerge after treatment with telaprevir and boceprevir. Perhaps the most important of these is R155K/T, the fitness of which can improve when it is associated with another RAV: V36M. The RAVs most frequently associated with second-generation protease inhibitors are, again, R155K and D168A/V/E/T. In the OPTIMIST study, of 26 patients treated with SOF+SMV who received 12 weeks of treatment without RBV, 16% failed treatment, 14 were sequenced, with NS3A RAVs detected in 11 at the time of treatment failure, and none showed SOF RAVs.
- ∘
NS5A RAVs: these are probably the most important problem facing therapists at present. Although NS5A inhibitors have pangenotypic coverage, each is associated with a variable specific activity and has a relatively low genetic barrier. The detection rate of NS5A RAVs in different direct population sequencing studies is 0.3%–3.5%.21 There are multiple potential regional variants that can induce resistance to NS5A inhibitors. Unlike NS3 RAVs, NS5A RAVs persist over time, and up to 85% are present 1–2 years after treatment completion,14,22 and may have an impact on the efficacy of future rescue treatments. Most RAVs are cross-linked, in other words, resistance to an NS5A inhibitor drug usually determines resistance to other drugs in the same group. In the case of DCV, the most frequently found are L31V/M and Y93H/N in genotype 1b, and Q30H/S in genotype 4.23 OBV RAVs are detected at positions 28, 30, 58 and 93, almost exclusively in genotype 1a infections. LDV RAVs with a greater influence on achieving SVR occur at Q30E/R, L31M, Y93C/H/N in genotype 1a and at Y93H in genotype 1b. The latter is the most common. It is found in 3.8%–14.1% of patients, and induces medium–high resistance to drugs acting on NS5A. This variant is more common in European (15%) than American (9.3%) patients.21 In a study of 2144 patients treated with LDV/SOF, of the 2.4% that presented virological failure, 74% had RAVs following failure, and most were genotype 1a.24 The combination of asunaprevir and DVC in patients with genotype 1a is currently not recommended in Europe and the USA due to the impact of NS5A RAVs on the SVR rate. The regimen is not recommended in patients with genotype 1b and baseline RAVs at L31 either because of its contribution to therapeutic failure.25 In another similar multicentre, phase III study in genotype 1b patients treated with ASN and DVC, NS3-D168 RAVs and NS5A-Y93 RAVs were detected in 75% of patients, with the potential impact on treatment response.26
- ∘
NS5B RAVs: these RAVs usually persist for a considerable time, and have been found up to 96 weeks after completion of treatment in 85% of NS5A patients, which is an indication of the greater viral fitness of these variants. The S282T variant is the only mutation associated with a decrease in susceptibility to nucleotide inhibitors, of which SOF is the paradigmatic molecule. Its prevalence is minimal, probably because its low fitness allows it to be rapidly replaced by the wild type virus,27 and it is unrelated to other NS5B RAVs.28 In the Electron study of SOF as monotherapy, the S282T RAV was detected in a patient infected with genotype 2 who relapsed at week 4 post-treatment.29 So far, this RAV has only been detected in 4 patients. The NS5B RAVs that confer resistance to non-nucleoside NS5B inhibitors (such as DSV or tegobuvir) are far more common than those affecting nucleotide inhibitors, and are linked to viral resistance and breakthrough. In general, they are found more frequently in genotype 1a than genotype 1b patients,30 although the C316N variant is observed more frequently in genotype 1b.31
- ∘
The most important impact of RAVs involves their capacity to reduce viral susceptibility to antiviral drugs. The only clinically relevant RAVs appear to be those that arise after DAA failure, and of these, those that appear in the NS5A region currently have the greatest impact. In a multicentre study analysing RAVs in 165 patients with pre-treatment failure from 33 Spanish hospitals using next generation sequencing with subtype-specific primers, resistance-associated RAVs were observed in 84.2% of cases.32 The presence of RAVs is not the only factor involved in therapeutic failure, and most authors agree that the association of RAVs with negative prognostic factors, such as viral genotype (1a, 3), the presence of cirrhosis, or shorter treatment regimens, has a greater impact on viral response. According to the ION-3 study, 24-week re-treatment regimens in patients with no RAVs who had received previous treatment for 8 weeks were associated with a very low SVR rate (below 50%) if they had developed 2 or more NS5A RAVs.33
Diagnostic methodsThe ability to detect RAVs depends mainly on the analytical method used. Traditionally, RAVs are detected using population sequencing (direct sequencing, PCR). This method can detect variants that represent at least 10% of the viral population.34 More sensitive techniques, such as clonal sequencing (plasmid vector cloning using PCR followed by direct sequencing), can detect variants that can account for only 1% of the entire viral population; however, the recent development of even more sensitive methods, such as next generation sequencing can detect variants that account for less than 0.5%–1% of the total sample.35 These techniques can also detect infection by more than 1 subtype simultaneously, which has obvious diagnostic and therapeutic implications. Next generation sequencing can also identify compensatory resistances and mutations in the same genome, together with recombinant viruses, subtypes and mixed infections.36,37 It can also determine the replicative capacity (fitness) of the virus.
Interpreting RAVs is not always straightforward, and will depend on the sequencing method used. The link between antiviral resistance and the RAVs detected by direct sequencing is clear, but many minority variants identified using highly sensitive techniques are not associated with any clinical phenomenon and have no impact on the choice of antiviral treatment. This is why RAVs should be interpreted by multidisciplinary groups formed of experts in the fields involved. A study on patients retreated with LDV-SOF noted that of all patients with RAVs detected by deep sequencing, only 60% of those receiving regimens that included RBV achieved SVR, whereas 100% of patients without RAVs achieved SVR.33 These tests are currently available in the USA, but they are expensive. They are not commercially available in Europe, but different research laboratories have experience in the analysis of NS3, NS5A and NS5B sequences. Most experts seem to agree on the need to perform resistance tests on all patients who fail DAA therapy in order to guide rescue therapy. This highlights the pressing need to market a standardised resistance test for these patients.
Re-treatment recommendations and strategiesBefore describing the treatment options in these patients, it is important to consider certain preliminary, pre-treatment aspects:
- -
Importance of correct genotyping in these patients.
- -
Assess the patient's clinical context, comorbidities, and therefore the urgency of re-treatment at this time. In non-cirrhotic genotype 1 patients with failure of NS5A inhibitors, the current AASLD guidelines recommend delaying re-treatment until new drugs have been developed.
- -
Analyse the previous treatment conditions, whether or not RBV was used, the duration of the treatment, possible non-compliance, and possible interactions with concomitant medication (proton pump inhibitors, for example, are frequently used).
- -
Consider the possibility that treatment failed due to reinfection (especially in certain groups, intravenous drug users, prison inmates, etc.), superinfection, or genetic recombination processes. Although these are not addressed in this review, they must be taken into account.
- -
Test for RAVs after failure of DAA therapy: this is a logical recommendation. Although complete sequencing of the viral genome is not available in all hospitals, it is important to consider taking baseline samples from all patients and storing them at −70°C in case the patient requires a full virological analysis if treatment fails. This sample can then be compared to a post-treatment sample, making it possible to analyse any RAVs that could be involved.
- -
Re-treatment strategies should include SOF (high genetic barrier, RAVs which rapidly disappear when treatment has stopped) accompanied by 1–3 additional drugs (ideally with no cross-resistance to those previously used, as in the case of NS5A).
- -
Treatment with RBV should last 12 weeks, extending to 24 weeks in the most difficult to treat patients, or 24 weeks without RBV in patients in whom the drug is contraindicated. An analysis of 513 treatment-experienced and treatment-naive cirrhotic genotype 1 patients receiving SOF/LDV±RBV for 12 or 24 weeks, showed that RBV is particularly useful in patients with RAVs, and increases the possibility of achieving SVR by almost 10%.38
Although there is little experience in the re-treatment of patients with RAVs, some data from clinical trials and real practice are now available:
- -
Failure of SOF+SMV: although certain cross-resistance between NS3 and NS5A RAVs has been described, re-treatment options must include NS5A inhibitors. Possible options are SOF+LDV or SOF+DCV.39,40
- -
Failure of SOF+LDV: failure of this combination is usually due to the appearance NS5A RAVs, which occurred in 76% of failures of this treatment.41 No S282T RAVs have been reported in the literature. In a study with 41 patients who failed SOF+LDV, all were retreated with the same regimen for 24 weeks. Thirty of these were treated with an 8-week regimen, with 80% achieving SVR vs only 46% of those previously treated with a 12-week regimen. Only 60% of patients with NS5A RAVs achieved SVR vs 100% of those without RAVs. The SVR rate in patients with 2 NS5A RAVs was 50%.41 In another study in genotype 1, patients previously treated with several NS5A inhibitors were retreated with SOF+3D. Although the sample size was small, 100% SVR was achieved in patients with NS5A RAVs (6 patients), the Y93H/N variant was particularly resistant to treatment (only 2 of the 6 patients with this RAV achieved SVR).42 A recent trial evaluated re-treatment with SOF/LDV+GS-9669+GS-9451 in patients failing SOF/LDV. The combination showed high antiviral efficacy, despite 1 case with a highly complex resistance pattern (L31M, Y93H, S282T, V321) which is currently untreatable.43
- -
Failures of DCV-containing regimens: data on SOF+DCV failure are similar to the SOF+LDV combination. The SOF+DCV combination has been evaluated in 152 treatment-naive and treatment-experienced genotype 3 patients. Relapse occurred in 9% of naive vs 14% of experienced patients, most of whom were cirrhotic. NS5A RAVs were observed, 6 at baseline and 10 after treatment, in all cases with relapse.44 A potential re-treatment regimen was recently explored in a pilot study45 evaluating the efficacy of SOF+SMV without RBV for 12 weeks in 15 patients who did not respond to DCV+PR (n=12) or DCV+ASV+PR (n=3). Response was achieved in 13 cases. The 2 patients presenting relapse were genotype 1a with cirrhosis who had baseline resistance to NS3 and NS5A (R155K, Q80K and V170I) and NS5A (M28T, L31M).25 The C-ISLE study presented its preliminary data at an AASLD conference. Study subjects were non-responders to previous treatments, including patients treated with SOF+DCV+RBV, who were retreated with SOF+GZP+EBV, achieving 100% SVR rates.46
- -
In another study, patients failing SOF+VPA were retreated with SOF+VPA+RBV for 24 weeks, achieving a high SVR rate in general, although SVR rates in genotype 3 patients with NS5A RAVs decreased to 77%.29
- -
Failure of 3D regimens: the efficacy of 3D regimens is very high, as high as that achieved with the SOF+LDV combination, although experience in re-treatment is equally scarce. In a study of more than 1000 treated patients, a deep sequencing study was performed on patients presenting virological failure. NS3–NS4 resistant variants were detected in 78% of genotype 1a and 57% of genotype 1b patients, most frequently in position D168. In total, 72% of patients with genotype 1a vs 29% of patients with genotype 1b showed variants resistant to NS5A. The number of patients with variants resistant to all 3 targets (NS3, NS5A and NS5B) was not published, but estimates based on study data show this to be over 70%.22 The following re-treatment options have been evaluated: the addition of INF and RBV to the 3D regimen, and the addition of SOF to the 3D regimen. However, the outcomes of these two strategies have yet to be confirmed. The QUARTZ-1 study analysed the re-treatment of patients failing 3D regimens (and other combinations) in 22 cases with a combination of paritaprevir, OBV, DSV, SOF with or without RBV for 12 or 24 weeks, achieving SVR in 20 cases. The 2 patients with relapse were cirrhotic, genotype 1a, and were treated for 12 weeks.47
Several clinical trials have published very promising preliminary data on potentially highly effective short-term re-treatment options. In a phase II study, treatment with SOF+VPA+voxilaprevir in cirrhotic and non-cirrhotic patients previously treated with DAAs found RAVs (15% NS3 RAVs, 31% NS5A RAVs, 27% mixed) in 73% of 48 patients undergoing sequencing. SVR was achieved in 100% (24/24) of patients treated with SOF+VPA+voxilaprevir without RBV, and in 96% (24/25) of those treated with RBV.48 Another combination currently undergoing a phase II trial is glecaprevir and pibrentasvir. In this case, 50 non-cirrhotic patients previously treated with DAAs have been studied, finding RAVs to NS3, NS5A and both, with SVR rates of 100%, 91% and 93%, respectively. An SVR rate of 100% was achieved in patients without RAVs.49
Clinical trials are the gold standard for evaluating intrinsic viral activity, resistance profiles, and adverse events, although interesting and promising approaches can be found in recent studies. A case in point is the recently published paper that combines an experimental phase with mathematical models in order to evaluate intrinsic viral activity and the theoretical appearance of resistance to double and triple antiviral combinations. These approaches will be very useful in designing drug combinations based on a more accurate antiviral profile and hypothetical resistances.50
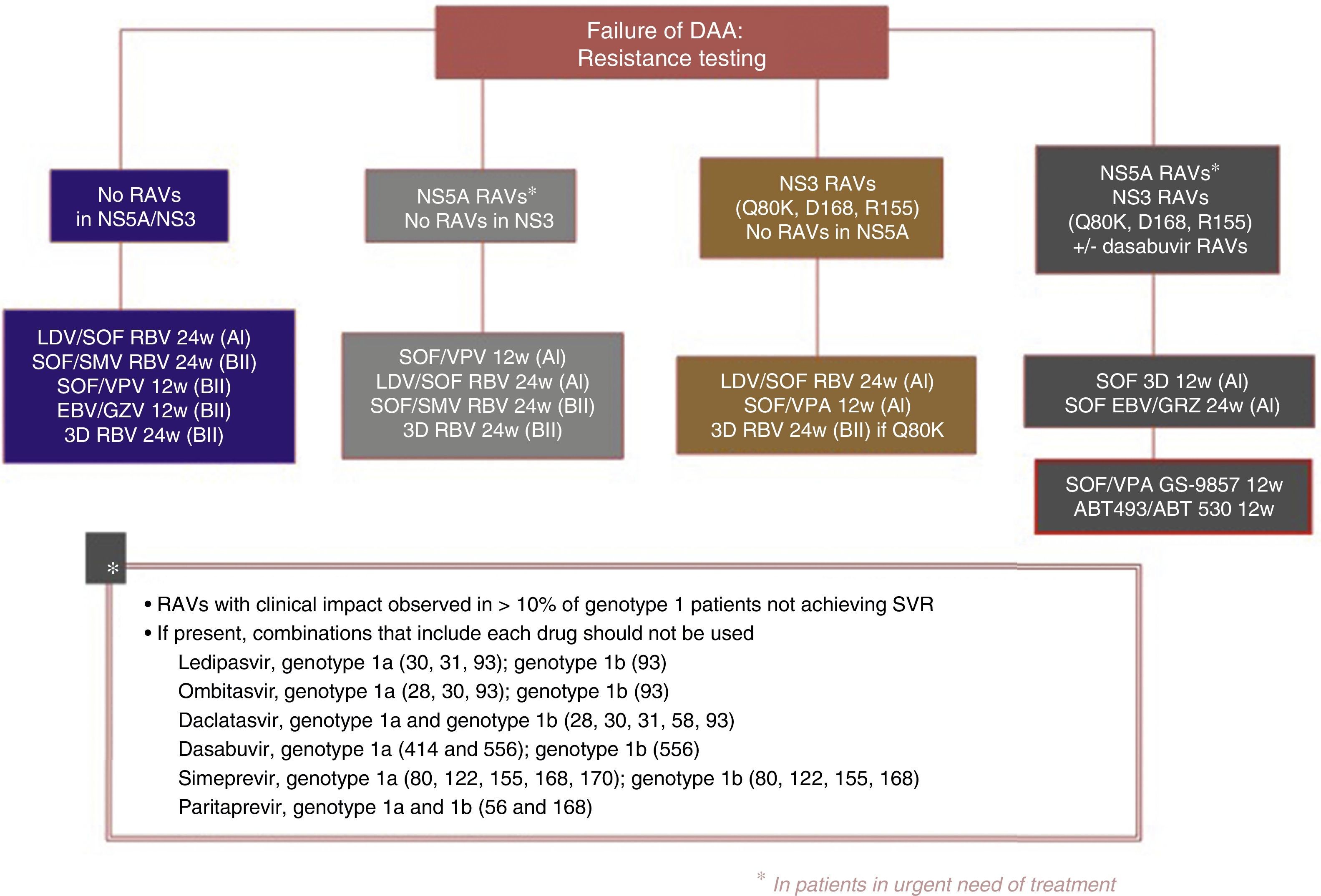
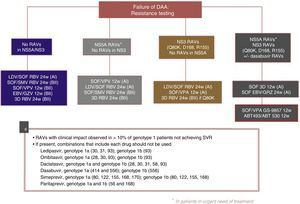
The following re-treatment regimens are recommended by the EASL51:
- •
Failure after PEG-IFN+RBV+(SMV, TPV, BOC): retreat with SOF+an NS5A inhibitor (LDV, DCV, VPA)+RBV for 12 weeks
- •
Failure after SOF alone, SOF with RBV or PEG-IFN+SOF+RBV regimens: in genotypes 1, 2, 3, 4, 5 or 6, retreat with SOF+an NS5A inhibitor (LDV, DCV, VPA)+RBV for 12 weeks in non-cirrhotic patients and 24 weeks in cirrhotic patients. Another option is SOF+SMV+RBV with the same recommendations regarding duration. In genotype 1 and 4 patients, 3D (PTVr, OBV, DSV) or 2D (PTVr, OBV) regimens can be used, according to genotype (1 or 4), or GZP/EBV.
- •
Failure after SOF+SMV: retreat with SOF+an NS5A inhibitor (LDV, DCV, VPA)+RBV for 12 weeks in non-cirrhotic patients, 24 weeks in cirrhotic patients.
- •
Failure following NS5A inhibitors: in genotypes 2, 3, 5 and 6, retreat with SOF+VPA+RBV for 24 weeks. In genotype 1: SOF+3D+RBV for 12/24 weeks, or SOF+GZP+EBV+RBV for 12/24 weeks, or SOF+DCV+SMV+RBV for 12/24 weeks. In genotype 4 SOF+2D+RBV for 12/24 weeks or SOF+GZP+EBV+RBV for 12/24 weeks or SOF+DCV+SMV+RBV for 12/24 weeks.
- ∘
General recommendations for baseline resistances studies cannot be made at this time. However:
- ∘
In genotype 1a with cirrhosis, Q80K polymorphism must be determined prior to the start of therapy or re-treatment with SMV-based regimens (A1)
- ∘
In cirrhotic, genotype 1a, null responders, the presence of EBV-resistant RAVs in NS5A must be ruled out before using the EBV/GZV without RBV combination for 12 weeks (A1)
- ∘
Resistance testing should be used to guide treatment changes in patients who cannot wait for new treatment regimens to be developed. If urgent re-treatment is not imperative, it is best to wait until new combinations become available (A1)
- ∘
If resistance testing is not available, re-treatment strategies must include RBV and extended regimens (A2)
Fig. 3 shows the recommendations contained in the most recent consensus document on re-treatment options and the combinations to be avoided with each specific RAV identified, published by the Spanish Association for the Study of the Liver.53
Recommended re-treatment regimens, according to the Spanish Association for the Study of the Liver.52
ABT 493: glecaprevir; ABT 530: pibrentasvir; DAAs: direct antiviral agents; EBV: elbasvir; GZV: grazoprevir; LDV: ledipasvir; RAVs: resistance-associated variants; RBV: ribavirin; SOF: sofosbuvir; SVR: sustained virologic response; SMV: simeprevir; VPA, velpatasvir.
In this age of DAAs that cure most patients, we need to turn our attention to the few, yet persistent, treatment failures.
These failures are due to both virological and non-virological factors, in which multiple agents each play a part.
We are improving our understanding of the behaviour and significance of RAVs, which are partly responsible for loss of susceptibility to antiviral agents. We emphasise the need to obtain baseline samples from all patients before the start of treatment. In this way, the determination and comparison of baseline and post-treatment RAVs in the event of DAA failure will guide the re-treatment regimen. These samples should be sent to specialised centres, and the results analysed by experts.
An understanding of RAVs coupled with the new knowledge brought to light in the latest clinical trials can provide us with effective re-treatment options for our patients.
Conflicts of interestJavier Crespo has received funding from AbbVie, BMS, Gilead, Janssen and MSD.
The remaining authors have no conflicts of interest to declare.
Please cite this article as: Llerena S, Cabezas J, Iruzubieta P, Crespo J. Resistencias al virus de la hepatitis C. Implicaciones y posibilidades terapéuticas. Gastroenterol Hepatol. 2017;40:484–494.