Colorectal cancer (CRC) is one of the common malignant tumors in the world. Colonoscopy is the crucial examination technique in CRC screening programs for the early detection of precursor lesions, and treatment of early colorectal cancer, which can reduce the morbidity and mortality of CRC significantly. However, pooled polyp miss rates during colonoscopic examination are as high as 22%. Artificial intelligence (AI) provides a promising way to improve the colonoscopic adenoma detection rate (ADR). It might assist endoscopists in avoiding missing polyps and offer an accurate optical diagnosis of suspected lesions. Herein, we described some of the milestone studies in using AI for colonoscopy, and the future application directions of AI in improving colonoscopic ADR.
El cáncer colorrectal (CCR) es uno de los tumores malignos más comunes del mundo. La colonoscopia es la técnica de examen crucial en los programas de detección del CCR para la detección temprana de lesiones precursoras y el tratamiento del cáncer colorrectal precoz, pudiendo reducir su morbilidad y mortalidad de manera significativa. Sin embargo, las tasas de pérdida de pólipos agrupadas durante el examen colonoscópico son tan altas como el 22%. La inteligencia artificial (IA) proporciona una forma prometedora de mejorar la tasa de detección de adenomas colonoscópicos (ADR). Podría ayudar a los endoscopistas a evitar la pérdida de pólipos y ofrecer un diagnóstico óptico preciso de las lesiones sospechosas. En este documento, revisamos algunos de los principales estudios sobre el uso de IA en colonoscopia y las perspectivas futuras en la mejora de la ADR colonoscópica.
Colorectal cancer (CRC) is one of the most common malignant tumors all over the world. According to global cancer statistics 2021, it ranks third in incidence and second in mortality.1 CRC arises from the epithelial cells of the lumen, and multiple disease processes, such as polyps, adenomas, and intraepithelial neoplasia, can participate in the transition from ordinary to advanced malignant mucous membranes.2 If screening can occur during this period, early detection may significantly decrease CRC mortality and morbidity. Additionally, the ADR is an independent indicator of interval colorectal cancer.3,4 The incidence of post-colonoscopy CRC and CRC-related mortality has been shown reversely associated with ADR during colonoscopy. There is evidence that for every 1.0% increase in ADR, the probability of interval CRC decreases by 3.0%.4,5
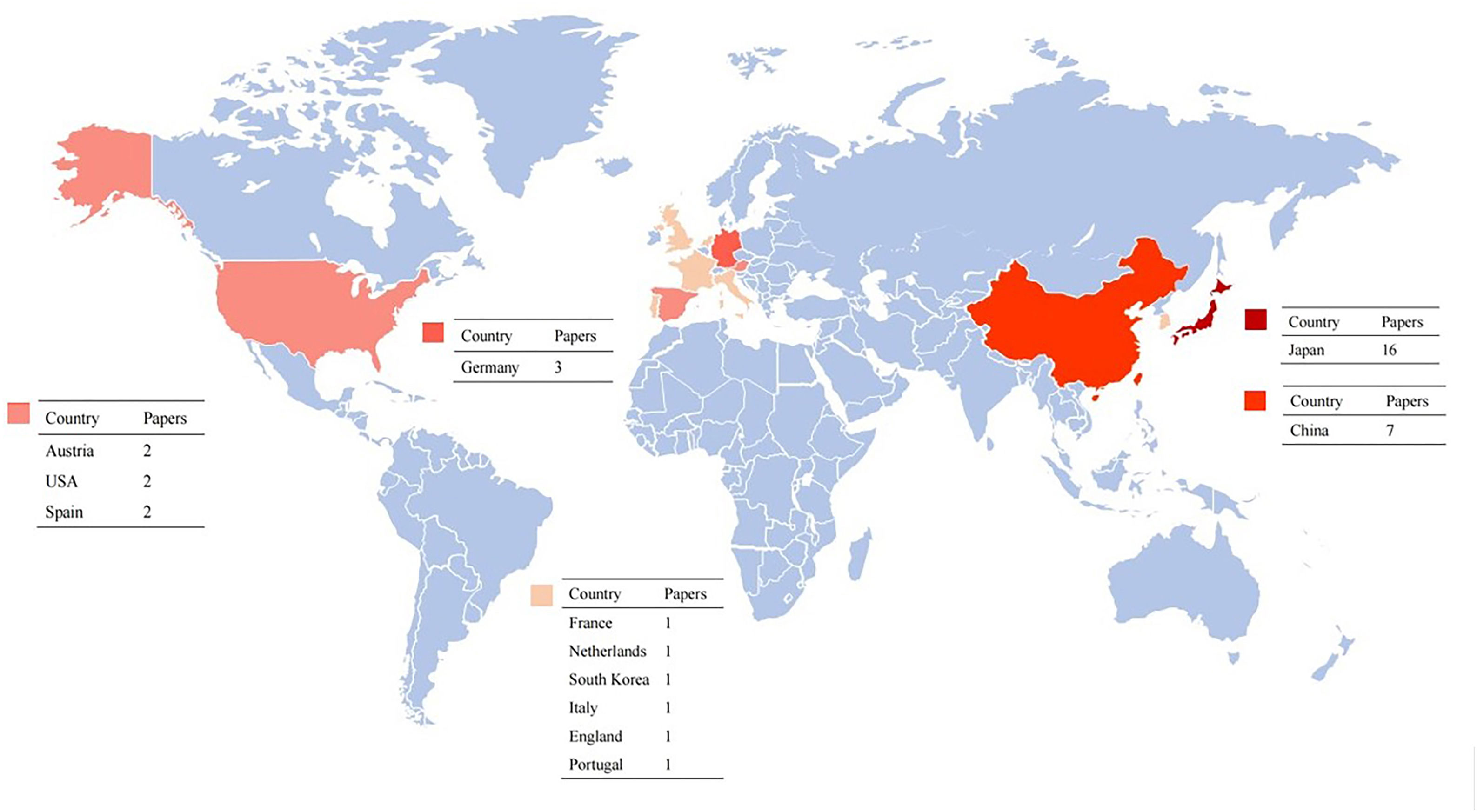
However, colonic polyps can be missed, with missing rates recorded up to 27%.6,7 Several factors were put forward to understand the reasons by which polyps may be missed, including differences in tracking patterns and skill level, the distraction caused by fatigue, the distraction caused by emotional factors, “change blindness,” and “inattentional blindness”.8–11 Besides, the analysis time must be quick for efficiency, and there should be no significant delay, leading to the lesions displayed on video monitors not recognized by endoscopists correctly. Furthermore, flat and depressed lesions are not only difficult to identify but are also more likely to present with advanced histopathology, whereas sessile serrated lesions show endoscopic characteristics such as mucus capping, unclear boundaries, and pale color. Because of these characteristics, they are more difficult to distinguish from background mucosa than typical adenomas.12 Artificial intelligence (AI) can improve medical diagnosis and treatment quality, overcome human errors and draw tremendous attention.13–17 To understand the current state of research and academic hotspots, we conducted this review (Fig. 1, Table 1).
Characteristics of studies on colorectal polyp detection and histology prediction by artificial intelligence.
| Author | Year | Study type | Country or region | Polyp no. | Patient no. | No. of polyp images | Imaging modality | Computer-aided model | Real-time | Video | Diminutive polyps involved |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tischendorf JJ39 | 2010 | Retrospective | Germany | 209 | 128 | NA | NBI | Region growing algorithm | N | N | N |
| Takemura Y40 | 2010 | Retrospective | Japan | NA | NA | 134 | Magnifying endoscopy | Watershed algorithm | N | N | N |
| Takemura Y41 | 2012 | Retrospective | Japan | 371 | NA | NA | NBI | Support vector machine (SVM) | N | N | N |
| André B58 | 2012 | Retrospective | France | 135 | 71 | NA | Confocal laser endomicroscopy | Retrieval-based software classification | N | Y | N |
| Kudo SE49 | 2014 | Retrospective | Japan | 514 | 455 | NA | Magnifying chromoendoscopy | CNN model | N | N | Y |
| Mori Y53 | 2015 | Retrospective | Japan | 176 | 152 | NA | Endocytoscopy, NBI | Support vector machine (SVM) | N | Y | N |
| Kuiper T59 | 2015 | Retrospective | Netherlands | 207 | 87 | NA | Laser-induced fluorscene spectroscopy | WavSTAT | N | N | Y |
| Fernández-Esparrach G24 | 2016 | Retrospective | Spain | 31 | NA | 612 | WLI | Window Median Depth of Valleys (WM-DOVA maps) | N | Y | N |
| Kominami Y42 | 2016 | Prospective | Japan | 118 | 41 | NA | NBI | Support vector machine (SVM) | Y | Y | Y |
| Mori Y54 | 2016 | Retrospective | Japan | 205 | 123 | NA | Endocytoscopy, NBI | Support vector machine (SVM) | N | N | Y |
| Misawa M56 | 2016 | Retrospective | Japan | 85 | NA | 1079 | Endocytoscopy, NBI | Support vector machine (SVM) | N | Y | N |
| Rath T60 | 2016 | Prospective | Germany | 137 | 27 | NA | Laser-induced fluorscene spectroscopy | WavSTAT4 | Y | Y | Y |
| Komeda Y37 | 2017 | Retrospective | Japan | NA | NA | 1200 | WLI, NBI, chromoendoscopy | CNN model | N | N | N |
| Zhang R48 | 2017 | Retrospective | China | 215 | NA | 826 | NBI, WLI | Support vector machine (SVM) | N | N | N |
| Takeda K55 | 2017 | Retrospective | Japan | 238 | 242 | 5543 | Endocytoscopy | Support vector machine (SVM) | N | N | N |
| Misawa M26 | 2018 | Retrospective | Japan | 155 | 73 | NA | WLI | CNN model | N | Y | N |
| Urban G27 | 2018 | Retrospective | United States | 4088 | NA | 8641 | WLI | CNN model | Y | Y | Y |
| Wang P28 | 2018 | Retrospective | China | 3634 | 1290 | 3634 | WLI | CNN model | Y | Y | Y |
| Mori Y57 | 2018 | Prospective | Japan | 466 | 325 | NA | Endocytoscopy, NBI | Support vector machine (SVM) | Y | Y | Y |
| Yamada M29 | 2019 | Retrospective | Japan | 752 | NA | 4887 | WLI | CNN model | Y | Y | Y |
| Wang P30 | 2019 | Prospective | China | 767 | 1058 | NA | WLI | CNN model | Y | Y | Y |
| Su JR31 | 2019 | Prospective | China | 273 | 659 | NA | WLI | Deep convolutional neural network (DCNN) models | Y | Y | Y |
| Sánchez-Montes C38 | 2019 | Retrospective | Spain | 225 | NA | 225 | WLI | Support vector machines (SVM) | N | N | Y |
| Min M51 | 2019 | Retrospective | China | 181 | 91 | NA | LCI | Gaussian mixture model (GMM) | N | N | N |
| Jin EH25 | 2020 | Retrospective | South Korea | 300 | NA | 300 | NBI | CNN model | N | N | Y |
| Liu WN32 | 2020 | Prospective | China | 734 | 1026 | NA | WLI | CNN model | Y | Y | N |
| Wang P33 | 2020 | Prospective | China | NA | 369 | NA | WLI | Deep convolutional neural network (DCNN) model | Y | Y | Y |
| Ozawa T61 | 2020 | Retrospective | Japan | 4752 | 3021 | 16418 | NBI, WLI | CNN model | N | N | Y |
N=No; Y=Yes; NA=not available; NBI=narrow-band imaging; WLI=white-light imaging; LCI=Linked-color imaging.
In mass media, AI is a term for machine mediated automation, intended to substitute human cognitive processes. Contemporary AI should be evaluated as complex algorithms, which can complete tasks without any clear guidance. This expertise is also known as machine learning (ML). ML models can extract and transform characteristics through different data sets and realize self-learning classification and prediction. One of the most popular uses of ML is the classification of objects such as lesions into specific classes based on input features obtained from segmented objects. In other words, the task of ML is to determine optimal boundaries based on the input features for separating classes. In this use, an ML algorithm is often called a classifier. “Deep learning (DL),” which means that several algorithms are merged into complex layers, is a superior type of ML. DL directly uses images as input (uses pixel values in images directly instead of features calculated from segmented objects as input information), does not require feature calculation or object segmentation. Because DL can avoid errors caused by inaccurate feature calculation and segmentation, which often occur for subtle or complex objects, the performance of DL is generally higher than ML. “Artificial neural networks (ANN)” or “convolutional neural networks (CNN)” are the common terminology used for AI. ANNs and CNNs with image input do the entire process from input images to the final classification. However, there are some differences between CNNs and ANNs. For example, CNNs require a large number of training images because of a large number of parameters in the model, whereas ANNs require a minimal number of training images. These interconnected algorithms can classify data features that cannot be completed by a separate algorithm on their own, exponentially boosting algorithmic learning ability. Most AI systems are based on DL methodology in contemporary times (Table 2).
The main AI systems.
| Name | R&D Companya | Year | Application |
|---|---|---|---|
| WavSTAT4 Optical Biopsy System | SpectraScience | 2016 | Real-time diagnosis of pre-cancerous colon polyps |
| Real-Time Automatic Polyp Detection System | Shanghai Wision AI Co., Ltd. | 2016 | Real-time auxiliary diagnosis |
| Endoscopic Video System (EVIS LUCERA ELITE CV-290/CLV-290SL) | Olympus Corporation | 2017 | Real-time monitoring of video images |
| ENDOANGEL | Wuhan EndoAngel Medical Technology Co., Ltd. | 2018 | Monitoring of video images and assisting physicians in prompting suspicious lesions in real-time. |
| The CADe system of polyps | Henan Xuanweitang Medical Information Technology Co., LTD., Zhengzhou City, Henan Province, China (Former name: Henan Tongyu Medical Technology Co., Ltd. | 2019 | Quality monitoring and auxiliary diagnosis |
| GI Genius™ Intelligent Endoscopy Module | Medtronic | 2019 | Detect colorectal polyps of various sizes, shapes, and morphologies |
| CAD EYE | FUJIFILM Europe GmbH | 2020 | Real-time detection of colonic polyps and real-time colon polyp characterization |
| ENDO-AID (EVIS X1) | Olympus Corporation | 2020 | Real-time display of automatically detected suspicious lesions |
The development of AI has gone through three waves. The first two waves happened in the 70s and 90s, respectively. The application of AI in different industries had not reached satisfactory results due to the algorithm's limitations at that time. With DL technology installed in 2006,18 AI ushered the third wave, allowing CNN to be widely used in image recognition.
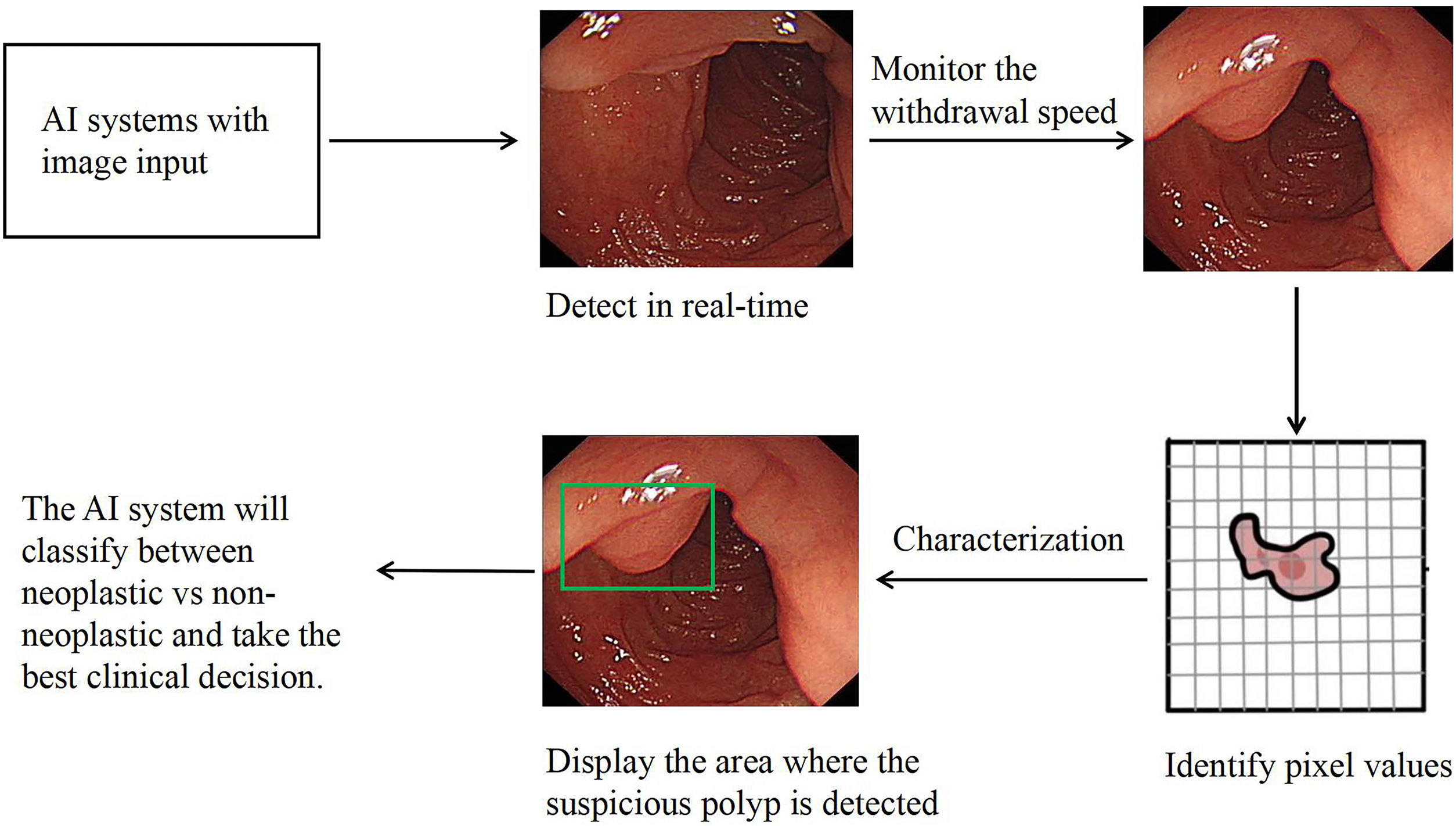
Computer-aided diagnosis (CAD) is growing into a common means of fixing the human error. Computer-aided detection and characterization of colorectal polyps are gaining popularity these days. For a long time, researchers have been researching the concept and use of CAD technology for colonoscopy.19,20 AI plays essential roles in colonoscopy practice, including computer-aided detection (CADe) and computer-aided characterization (CADx). By displaying the appearance and position of polyps during colonoscopy in real-time, CAD theoretically has the potential to draw the endoscopist's attention to polyps that are visible on the monitor but maybe overlooked visually. This, in turn, will result in a higher ADR. Additionally, by providing the predicted pathology or endoscopic classification of any identified polyps, CADx contributes to the accurate characterization of colorectal polyps (Fig. 2).
Diagnostic performance of AI in polyp characterizationThe application of AI in colonoscopy can improve the quality of colonoscopy.13,20 AI can automatically monitor the real-time withdrawal speed of colonoscopy and remind the endoscopists to control the withdrawal speed within a safe range during colonoscopy. The above characteristic can lead to an increase in ADR and the average number of polyps found in colonoscopy patients when the withdrawal time was more than 6min.21,22 The incidence of colorectal cancer was significantly lower in patients who had longer withdrawal time.23 CAD also improves ADR by generating predicted pathology or endoscopic classification, resulting in a substantial decrease in unnecessary polypectomies for non-neoplastic polyps (Table 3). To denote the specified area of an image containing a polyp, Fernandez-Esparrach and colleagues used a window median depth of valleys accumulation (WM-DOVA) energy maps system.24 This system modeled polyps as protrusions in the mucosa and defined their boundaries. It is noteworthy that the WM-DOVA method was especially beneficial for the small flat (Paris 0–II) lesions and was not negatively affected by bowel preparation. Identification mistakes induced by lateral observations into polyps and other structures, including colonic folds and blood vessels, were among the model's shortcomings. Eun Hyo Jin et al. developed a CNN that significantly increases the accuracy of evaluating diminutive colorectal polyps (as adenomatous and hyperplastic) and reduces the time of diagnosis by endoscopists, found that AI is most effective in assisting novices than experts.25
Artificial intelligence pros and cons.
| Pros | Cons |
|---|---|
| Reduce errors caused by fatigue | May make endoscopy more costly in the initial stages of clinical use |
| Predict the pathology or endoscopic classification of polyps | AI-induced distraction, especially if the output was inaccurate |
| Real-time automatically monitor the withdrawal speed of colonoscopy | Over-reliance may cause endoscopists to be less skilled and meticulous |
| Decrease in unnecessary polypectomies | Biased datasets for ML may lead to unexpected results |
| More effective in assisting novices | Hacking attacks |
| Improve the quality of colonoscopy and ADR | Risk of loss of important data |
ADR: adenoma detection rate.
A critical objective of CADe in colonoscopy is to avoid missing polyps during colonoscope withdrawal. In 2016, Fernández-Esparrach et al. published an early study on automated polyp detection.24 They evaluated the CADe model using video recordings of 31 polyps and found a sensitivity and specificity of more than 70%. Following this study, Misawa et al. developed a real-time CADe algorithm and evaluated its effectiveness with 50 polyp videos and 85 non-polyp videos, achieving 90% and 63% sensitivity and specificity, respectively.26 Urban and colleagues also developed a CADe model. They reported an area under the curve of 0.991 (a metric in which values of 0.5 correspond to chance observation and values of 1.0 correspond to perfect accuracy) and an accuracy of 96%.27 Wang et al. also have developed a CADe model with over 90% sensitivity and specificity for video-based analysis.28 More recently, Yamada et al. trained their CADe using 1244 polypoid lesion images, 891 frames obtained from videos, and 2843 non-polypoid lesion images, achieving 97.3% sensitivity and 99.0% specificity. Interestingly, the sensitivity decreased to 74.6% when input video data into their CADe.29 Several prospective randomized control trials using CADe have recently been reported.30–32 These tests of CADe have all demonstrated considerably higher ADR. Wang et al. recently published a randomized trial using their CADe system. This study helps analyze the effectiveness of CADe systems since it assessed the adenoma miss rate, which is another valuable colonoscopy efficiency indicator. Adenoma miss rate in this study was substantially lower than without CADe (13.89% vs. 40.00%, P<0.001).33 Hence, CADe systems appear to increase ADR. Until recently, the study of Hassan C et al. supported the benefit when adding CADe to colonoscopy, argued for the independence between the additional benefit and the traditional features of colorectal neoplasia.34 In detail, CADe led to a significant increase in the ADR, regardless of whether the polyps were diminutive, small, or large, of those located in the proximal or the distal colon, and of those flat and polypoid. However, in their study, CADe had no meaningful effect on the efficiency of colonoscopy due to the similar withdrawal time. They also argued that specific studies are needed for both low- and high-detectors precisely because the relationship between baseline ADR and CADe benefit could not be properly assessed at present.
Computer-aided characterizationThe CADx is designed to predict the pathology of polyps. CADx could theoretically improve the precision and pathology evaluation of optical biopsy, thus reducing excessive resection of distal non-neoplastic polyps.35 It will also make it possible for novice endoscopists to adopt the technique of resect-and-discard strategy in clinical practice.36
The most basic and practicable modality of endoscopy diagnosis is white-light endoscopy (WLI). Hence, the CADx application to WLI would also be beneficial. Komeda et al. reported a CADx using DL for distinguishing adenomatous and non-adenomatous lesions.37 However, its accuracy of diagnosis was just 75.1%. Further tuning of the algorithm was then required. Sánchez-Montes et al. have developed a CADx for WLI to assess whether or not a lesion is dysplastic. Their accuracy was 91.1%.38 Nevertheless, CADx for WLI would not be as reliable as CADx for narrow-band imaging (NBI) or other image enhancement techniques. Further research is also essential to show the ability of CADx for WLI.
NBI is an image-enhanced technique that enables a comprehensive assessment of the surface vessel and the structural patterns. Tamai et al. investigated the use of CADx in early CRC depth diagnosis, achieving deep submucosal invasive cancer prediction in 82.8% lesions. Tischendorf and colleagues first tested the computer-aided classification of colorectal polyps with NBI magnification images and zoom endoscope.39 Their CADx algorithm was built on extracting vessel characteristics from magnified NBI pictures and then classifying these features as neoplastic or non-neoplastic. This study obtained a sensitivity of 90% and an accuracy of 70.2% in discriminating between neoplastic and non-neoplastic images compared to histopathology as the gold standard. The outcome, however, was inferior to human observers. Magnification imaging was used in many subsequent experiments in Japan. For example, to classify pit patterns, Takemura and colleagues developed image analysis tools, reached an accuracy of 98.5% for their machine algorithm.40 One significant restriction was the semi-automation of the system, specific images require a manual processing stage, and the decision took some minutes.
Further retrospective research of this group used a computer-based system based on the Hiroshima classification to characterize 371 polyps into neoplastic or non-neoplastic.41 The surface structure and microvessels identified by NBI are classified into A types (non-neoplastic) and B–C types (neoplastic). For diagnosis of neoplastic lesions (type B–C3), the software achieved an accuracy of 97.8%, a sensitivity of 97.8%, and a specificity of 97.9%. The techniques, however, also required experts to pick and manually extract regions of interest. Subsequently, the upgraded software was evaluated prospectively.42 It is worth noting that, in Japan, the research group at Hiroshima University played an important role in developing CAD models.43–47 Their CADx works in real-time and has been used for 93.2% precision in a real-time clinical trial for separating adenomas from non-neoplastic polyps.42
To overcome the limitation of small labeled datasets, Zhang and colleagues developed a CNN for polyp classification using the transfer learning principle.48 1930 pictures included in the endoscopy dataset (1104 non-polyps, 263 hyperplastic polyps, and 563 adenomatous polyps). The images were taken from 215 polyps (65 hyperplastic and 150 adenomatous) and zoomed in with WLI and NBI images. The overall accuracy reached 85.9%, the sensitivity reached 87.6%, which was higher than the results of endoscopists measured on the same dataset.
Indigo carmine or crystal violet magnifying chromoendoscopy assists endoscopists in identifying the surface structure of colorectal polyps, leading to the high accuracy of lesion pathology predictions in the pit patterns diagnosis.49 CAD systems generally use one of two methods for pit pattern diagnoses: quantitative pit structure analysis or texture analysis of the whole endoscopic image. Takemura et al. utilized the former approach and achieved an overall accuracy of 98.5%.40 Although these approaches have shown outstanding diagnostic accuracy experimentally, up-to-date, no further assessment has been performed.
Linked color imaging (LCI) system, a new endoscopic imaging modality, has been developed in recent years. It can produce clear and bright images by using short wavelength narrow-band laser light. This newly emerged modality can enhance the colors of lesions during a colonoscopy, emphasize vascular and surface structures and color differences while maintaining a bright vision, and make red areas appear redder and white areas appear whiter.50 Adenomatous and non-adenomatous polyps may be possible to be distinguished by color evaluation on LCI images. In 2019, Min et al. trained a CADe system based on LCI Images.51 This system can obtain the histology prediction of polyps by analyzing the lesions’ colors, achieved an accuracy of 78.4%, a sensitivity of 83.3%, a specificity of 70.1%. The results indicated that a novel CADe system based on LCI could be a rapid and powerful decision-making tool for endoscopists. However, further research is still needed.
Endocytoscopy is a microscopic imaging modality in vivo contact. This helps endoscopists during colonoscopy to obtain real-time cellular images with 500-fold magnification power, providing the ultra-magnification capability to visualize nuclei.52 This device is thought to be ideal for CAD, because it still offers focused, fixed-size images that lead to a more rigorous but easier CAD image analysis. A Japanese research group has conducted an in-depth study of endocytoscopy with CAD. Mori and colleagues tested an endocytoscope CAD system (EC-CAD) in a pilot study.53 The EC-CAD automatically extracted characteristics from nuclei, and the pathological classification can be predicted within 0.3s. In terms of identifying neoplastic polyps, the sensitivity reached 92.0%, and the accuracy reached 89.2%. This performance was similar to that of experts. The specificity was 79.5%, and there were no significant differences between the experts or trainees. The same group has tested a second-generation EC-CAD with web-based image analysis.54 It extracted 296 features and classified polyps as nonneoplastic, adenoma, or invasive cancer and provided a diagnostic output in 0.2s. Its accuracy was 89%, and with a sensitivity of 89%, a specificity of 88%. The outcome again equivalent to the performance of experts and is substantially higher than that of non-experts. This system was subsequently utilized to discriminate between invasive cancer and adenomatous lesions, providing a sensitivity of 89.4%, a 98.9% specificity, and an accuracy of 94.1% for 188 Images.55 Endoscopic treatment will not be curative in the presence of deep submucosal invasion. Therefore, this differentiation is critical in guiding therapy. They also developed a more user-friendly CAD system based on endocytoscopy combined with NBI (EC-NBI), which overcomes the constraint of pre-staining lesions with dyes, which is necessary with traditional endocytoscopy.56 The system is mainly used to analyze the microvessels on the surface of polyps, with an overall accuracy rate of 90.0%. Recently, to evaluate the performance of real-time CAD with endocytoscopes providing microvascular and cellular visualization of colorectal polyps after application of the NBI, Mori et al. conducted a prospective study.57 Overall, 466 diminutive polyps (including 250 rectosigmoids) from 325 patients were assessed by CAD, with a pathologic prediction rate of 98.1%.
Another in vivo contact microscopic imaging method, confocal laser endomicroscopy, enables endoscopists to get real-time cellular images with 1000-fold magnification power during colonoscopy. There have been various researches to evaluate confocal endomicroscopy. 89.6% of the accuracy of the adenoma differentiation from the non-neoplastic polyps was reported in studies performed by Andre et al.58
Laser-induced autofluorescence spectroscopy uses an optical fiber integrated into the biopsy forceps to enable optical biopsy of colorectal polyps. The fiber emits laser light, and the tissue absorbs it. CAD then analyses the polyp's resultant autofluorescence signal, offering an in-vivo prediction of neoplasia. Kuiper et al. and Rath et al. published a CAD system with laser-induced autofluorescence spectroscopy.59,60 Both research groups evaluated this CAD system's performance for predicting polyp pathology (namely neoplastic or nonneoplastic) in real-time. The system is integrated into standard biopsy forceps. Endoscopists can then use these forceps to detect diminutive polyps, obtain pathological results predicted by CAD, and resect them later with the same biopsy forceps as needed.
Simultaneous usage of computer-aided detection and computer-aided characterizationThe combination of CADe and CADx is considered to be the most desirable. A Japanese research group presented a technique they developed based on the DL algorithm, which can simultaneously detect and predict polyp pathology. The system is designed to detect polyps in WLI, the endoscopic image obtained by photographing with the endoscope's tip in contact with the polyp.15 Ozawa et al. reported a similar system that can detect and characterize targets simultaneously.61 Their system, trained on 16,418 images from 4752 polyps, could predict one of five histopathological categories (hyperplastic, sessile serrated lesions, adenoma, cancer, and others). Their system had a sensitivity of 92% for WLI detection of colorectal polyps and the accuracy of characterizing reached 83%. These technologies are encouraging since both CADe and CADx are critical components of clinical colonoscopy.
Current limitations and future directions of AI in improving ADRCurrently, the use of AI in colonoscopy still carries such shortcoming, such as AI-induced distraction, over-reliance, biased datasets for ML, and hacking (Table 3). Endoscopists with CAD must pay heed while carrying out their examination of the output of CADe and/or CADx. This might distract endoscopists’ attention, resulting in missing or mischaracterizing polyps, especially if the output was inaccurate. Because AI provides a sense of security, over-reliance on AI may cause the next generation of endoscopists to be less skilled and meticulous.
Although the current findings appear encouraging, the data supporting CAD combined with colonoscopy is still finite. Furthermore, because the majority of the studies are retrospective, there may be considerable selection bias that leads to conclusions in favor of CAD. Some well-designed prospective trials were statistically more trustworthy than others because they eliminated the possibility of lesion selection bias, accounted for missing data, evaluated the success rate of data interpretation and the accuracy of CAD systems. However, the number of such trials is limited.
There are currently no AI systems for a colonoscopy that incorporate data from different countries since ADR may be influenced by several variables like genetics, lifestyles, diet, and behavior, and the incidence of colonic polyps/adenomas, polyp morphology, and the quality of bowel preparation can vary considerably between different countries. Similarly, differences in endoscopy technology between endoscopy manufacturers in different regions of the world may significantly affect the performance of the AI if the training datasets do not contain a full range of data. Small and unrepresentative datasets may lead to unexpected results.
AI may make mistakes in diagnosing and interpreting lesions. Such unintended, potentially negative effects can result in possible ethical and legal challenges. Therefore, to address the relevant conflicts underlying the adoption of AI, medical malpractice insurance may be one aspect of efforts. It needs to be clear about coverage of medical malpractice insurance when healthcare decisions are made in part by AI. This can make up as much as possible for the personal economic loss in the scenario of mistakes in the diagnosis and interpretation of lesions induced by AI. The government also needs to clarify the legally enforceable circumstances to provide further protection for individuals.
On the other hand, who should be held legally responsible for errors caused by the AI system? First, the government should check the performance of the AI system, set standards for it, and perform regular quality monitoring. Second, for clinical use, Research and development company (R&D company) must justify their analysis, particularly if there are concerns about unreliable predictions. Otherwise, for major medical accidents caused by the AI system, R&D companies may not avoid legal liabilities. In this regard, the future AI in colonoscopy should be international verification before being used on a global scale. The AI system should use images that adequately represent the target patient population, with a balanced ratio of high-quality and low-quality images, polyp and non-polyp images, and neoplastic non-neoplastic images to achieve good training results. Larger numbers of training images will contribute to better diagnosis accuracy, while the minimum amount to hit the learning plateau is still in an exploratory process.
Although many automated polyp detection systems have been built in the past decade, evidence for the capacity of this technology to locate and trace polyps during live colonoscopy is relatively lacking. There are insufficient data to determine whether sessile serrated or relatively flat and depressed lesions (Paris 0–II) are effectively identified. A high rate of false-positive results would complicate the endoscopists’ picture perception task and reduce colonoscopy quality. In the short term, diagnoses of endoscopists may be adversely affected by incorrect predictions.
An automated polyp detection device must have high sensitivity and specificity, appropriate real-time standard processing time, and an on-screen alerting system to be clinically applicable. Additionally, the delay should be as low as practicable, from collecting endoscopic image frames to producing the results examined. Owing to the higher instability of the colonoscope in the caecum and ascending colon, thus reducing the visual fields, AI may not have helped the endoscopist detect more adenomas in these areas. Future studies may focus on improving ADR in the caecum and ascending colon. The rise in overall ADRs may potentially lead to a decreased risk of interval CRC. More research can explore the role of CADe in reducing interval cancer, the principal target of any colonoscopy screening.
The polyp detection rate will further increase with the usage of modern technology. The vast majority of polyps detected during colonoscopy are diminutive (1–5mm) or small (6–9mm), and diminutive polyps account for approximately 60% of all detected polyps.62 These lesions have a low risk of causing advanced pathology or cancer. Furthermore, diminutive hyperplastic polyps’ detection rate may also be improved significantly, leading to additional unnecessary polypectomies, further adding to the workload. In the future, more clinical studies should make the CADe system integrated into a CADx system to support detection and diagnosis, as far as disregard strategy to avoid excessive workload.57
ConclusionAI technologies provide great potential for colonoscopy, are undoubtedly an appealing choice for improving the quality and efficiency of colonoscopy and standardizing the practice of colonoscopy. Real-time feedback might be valuable for improving ADR, guiding clinical decision-making, and reducing variance in colonoscopy quality. Several relevant technologies and their supporting evidence are growing significantly. In the future, we will need to conduct rigorous, high-quality prospective clinical studies to accumulate evidence and demonstrate the effectiveness of CAD for colonoscopy. Additionally, a solid collaborative connection between endoscopists and computer scientists is required to break down technological barriers and overcome future challenges. This is crucial for broader implementation. The increasing industry participation, government incentive measures, and assistance could contribute to significant advances over the coming years. In addition, considerations regarding obtaining regulatory clearance for early clinical use are necessary. With these kinds of inadequacies properly addressed, CAD will eventually lead to a new colonoscopy practice generation.
Author contributionsStudy conception and design: Xiaowei Tang, Xian Zhou. Drafting of manuscript: Peiling Gan. Acquisition of data and critical revision: Peiling Li, Huifang Xia. Revision of manuscript, and final approval of manuscript: Xiaowei Tang.
Conflict of interestDr. Peiling Gan, Dr. Peiling Li, Dr. Huifang Xia, Xian Zhou, Dr. Xiaowei Tang declare that they have no conflict of interest.
This study is independent research funded by the following grants: Youth Foundation of Southwest Medical University (No. 0903-00031099), Doctoral research start-up funding project of Affiliated Hospital of Southwest Medical University(No. 16229), Cooperation Project of Southwest Medical University and Luzhou Government (No. 2019LZXNYDJ24).