The diagnosis of invasive fungal disease remains challenging. The development of the European Organization for Research and Treatment of Cancer/Mycoses Study Group criteria has helped standardise diagnosis, yet PCR is excluded, a result of limited standardisation and concurrent clinical validation. In 2006, the European Aspergillus PCR Initiative working party of International Society of Human and Animal Mycoses was formed with the aim of establishing a standard for PCR methodology for the diagnosis of invasive aspergillosis, to attain an accurate clinical utility of PCR and acceptance of PCR in future disease defining criteria. This manuscript will provide an overview of the standardisation process, before describing the potential samples available and the reasoning behind the choices made. It will summarise the key findings, provide the European Aspergillus PCR Initiative recommendations when testing whole blood and serum, and conclude with a synopsis of future European Aspergillus PCR Initiative perspectives and other key processes that will improve PCR based diagnosis.
El diagnóstico de la enfermedad fúngica invasora sigue siendo un reto. El desarrollo de los criterios del grupo de trabajo de la European Organization for Research and Treatment of Cancer/ Mycoses han ayudado a estandarizar el diagnóstico, sin embargo, la prueba de PCR (reacción en cadena de la polimerasa) se excluye por su limitada estandarización y su concurrente validación clínica. En 2006, se formó la Iniciativa Europea Aspergillus-PCR, un grupo de trabajo de la International Society of Human and Animal Mycoses que tenía como objetivo establecer una metodología estándar de la PCR para el diagnóstico de la aspergilosis invasora y conseguir la utilidad clínica precisa de esta prueba para definir los criterios de la enfermedad. Este estudio ofrece una visión general del proceso de estandarización, describe las posibles muestras disponibles y el razonamiento en el que se basaron las decisiones tomadas. Resume las principales conclusiones, ofrece las recomendaciones de la Iniciativa Europea de Aspergillus PCR en el análisis de de la sangre total y el suero, y concluye con una sinopsis de las futuras perspectivas europeas de la iniciativa Aspergillus y los otros procesos clave que mejorarán el diagnóstico basado en PCR.
Molecular diagnostics brings many challenges to labora-tories and clinicians. Orderly and systematic evaluation of analytical, laboratory and clinical validity and utility is required. Nowhere is this more evident than in the field of invasive fungal disease (IFD) diagnosis where traditional methods lack sensitivity to detect disease
The European Organization for Research and Treatment of Cancer/Mycoses Study Group (EORTC/MSG) consensus definitions use a combination of host, clinical and mycological criteria to define IFD for the purpose of clinical trials1. Whilst not designed for diagnosis of IFD in other clinical settings, they are used increasingly. A proven diagnosis requires culture or histological demonstration of infection from a sterile site and, although yeasts, dimorphic fungi, and the occasional mould can be diagnosed by blood culture, invasive sampling procedures are required for invasive aspergillosis (IA), usually in critically-ill patients and, as consequence, are seldom performed. The “less certain” diagnosis of probable IFD, relies on seeing “specific” radio-logical evidence of a disease, i.e. those that are consistent with an IFD, in combination with mycological evidence in the form of demonstrating the presence of circulating biomarkers (Galactomannan [GM] or β-D-glucan [BDG]) or culture, in a patient with host factors.
PCR was not included as mycological evidence in the EORTC/MSG criteria despite significant research being done, particularly for IA2. Consequently, the value of PCR in aiding diagnosis is often overlooked. Exclusion from the EORTC/MSG criteria is less related to unsatisfactory performance but due to the fact that little standardisation had been attempted and, until recently, commercial alternatives were not available. Meta-analyses of GM, BDG and Aspergillus PCR in a clinical setting show similar sensitivities and specificities (BDG: 77% (95% CI: 67–84)/ 85% (95% CI: 79–89); GM: 78% (95% CI: 61–89)/81% (95% CI:72–88); PCR: 75% (95% CI: 54–88)/87% (95% CI: 78–93)3–5. The numerous publications relating to Aspergillus PCR reflect the continued scientific interest. There is methodological diversity with respect to PCR, though independent metaanalysis shows its performance to be comparable to other biomarkers that use a standard commercial approach. Nevertheless, PCR provides similar pooled performance to commercial standardised antigen detection, suggesting that standardisation of an optimal PCR protocol could provide superior performance.
With this in mind, two national studies were conducted focussing on comparing PCR methodology for the detection of IFD, in particular Candida and Aspergillus PCR, as these are the most frequent causative agents for IFD6,7. The UK and Ireland fungal PCR consensus group found that Candida PCR methods provided comparable and satisfactory performance, achieving a reproducible detection limit of 10 organisms8. Performance of Aspergillus PCR methods was more variable, and even when two optimal protocols were assessed by multi-centre evaluation performance of one assay varied with both PCR platform and sample type, and interactions between oligonucleotides and human (deoxyri-bonucleic acid (DNA) were noted. Performance variability was also noted by the German/Austrian inter-laboratory comparison of PCR methods, and this occurred on both an intra and inter-laboratory scale9. These studies highlighted the need for standardisation, particularly if Fungal PCR was to gain widespread use outside of specialised molecular/ mycology centres.
In 2006, the European Aspergillus PCR Initiative (EAPCRI) working party of International Society of Human and Animal Mycoses was formed with the aim of establishing a standard for PCR methodology for the diagnosis of IA. With over 62 participating centres from 25 countries in four different continents (Europe, North and South America and Australia), the EAPCRI is now a global initiative with several publications to its name10,11. This article will review the process and findings of the standardisation process for Aspergillus PCR and attempt to describe the differences when using different sample types.
PCR standardisation - procedural overviewBefore implementing any test, it is essential to understand both the incidence and pathology of disease, and the impact on sample selection. These factors affect the amount and specific target within this specimen. On agreeing a sample type it is important to consider how best to evaluate current methods and understand the individual stages the testing process that may impact on assay efficiency. This requires the evaluation of relevant individual stages as well as of the entire process. In the initial stages of the evaluation, logistical and ethical issues mean it is unlikely that any samples will come directly from infected patients. The use of animal models for large scale multi-centre investigations is not possible due the number and volume of samples required. Consequently, simulated samples are usually used, and the Quality Control (QC) panel should contain burdens and targets representative of the clinical scenario, as well as both negative and positive samples. The sample matrix should be screened for contamination before use and, if clean, the panel should be developed using good aseptic and molecular techniques, if necessary using clean room facilities (e.g. laminar air flow) to prevent preparation-borne contamination12.
Once a QC panel is prepared, it should be evaluated to determine expediency prior to storage to minimise degradation. The panel should be distributed in a way that maintains the storage status and sample anonymity. Problems with distribution should be recorded, particularly if they affect the panel status and the individual recipient should relay this back to the distributor. The panel should be stored by the recipient as agreed until testing, using standard protocols for that centre, and meeting any pretest conditions set by the distributor (e.g. testing time, internal control or replicate testing). In addition to the results, technical details concerning the entire molecular process should be returned. Overall multi-centre results should be compared to the original expediency test to establish if the panel has withstood the storage and distribution process. Blinded meta-regression analysis of results to determine assay performance and, both positive and negative, associations with individual technical steps should be performed. Any findings will form the basis of technical recommendations to be conveyed to participants and assessed by the distribution of further anonymous panels.
On return of the results and prerequisite technical information, blinded analysis will be performed for centres following recommendations designed to improve performance and compared to previous panels or to centres not following the recommendations.
If performance associated with the recommendations is shown to be superior then these are further assessed using specimens derived from animal models of the specific disease, ahead of any multi-centre clinical evaluation using patient samples.
Standardisation of Aspergillus PCR - the needWith the first manuscripts describing the use of PCR to aid in the diagnosis of IA being published almost two decades ago, it is surely time to finally decide whether PCR is clinically useful or not. GM ELISA has been in use for a similar period, is widely used for screening, diagnosis and monitoring of thera-peutic response, and methodological standardisation has been achieved by a single manufacturer adopting a commercial QC of the entire manufacturing process. By contrast PCR tests are mostly “in house” tests. Clearly methodological standardisation is needed13. The only way to accurately determine the clinical utility of PCR is through a prospective multi-centre study of substantial size, and the potential role of PCR can be determined through standardisation which does not exclude commercial participation, providing a much needed additional test for inclusion into a diagnostic strategy. Once this occurs, PCR can take its place among the tests accepted for defining IFD.
The European Aspergillus PCR Initiative structureInitially, the EAPCRI (http://www.eapcri.eu) created two working groups with extensive experience in laboratory (laboratory working group) and clinical (clinical working group) diagnosis of IA. Membership to these groups initially relied on pairing clinicians and scientists from the same centre. Both working groups had a lead member who represented the working parties on the steering committee, along with the chair of the organisation, whose role was to direct the EAPCRI process. However, it quickly became apparent that the laboratory working group programme required expansion and so additional test centres were recruited to help develop and test the protocols required for standardisation. The amount of information generated was such that a statistical working party was formed to help analyse the results.
Potential specimensA literature review of the pathology of IA, the range of specimens tested, and how these could affect the manage-ment of patients was undertaken before any laboratory evaluation could take place.
Respiratory SamplesSince IA most commonly affects the lungs, a respiratory speci-men seems the obvious choice to permit early detection of the organism. However, sputum is of limited use, associated with higher levels of airway contamination and can be difficult to process, whereas bronchoalveolar lavage (BAL) has great clinical relevance but is a relatively invasive procedure. The performance of PCR in BAL specimens is comparable to those for GM, BDG and PCR performed on serum or blood. A meta-analysis of Aspergillus PCR when testing BAL specimens provided pooled sensitivity and specificity of 79% and 94%, similar to the sensitivities and specificities generated by respective meta-analyses of GM, BDG and Aspergillus PCR testing blood specimens3–5,14. However, these observations do not take into account the temporal relationships of positivity, nor which is the earliest marker of disease, which is important as the prognosis is dramatically improved by early treatment led by an earlier diagnosis15. Aspergillus was detected in blood samples of cases of IPA at a later stage than was found for BAL16. However, obtaining BAL specimens may not be possible for a variety of reasons, including thrombocytopenia. Moreover, Aspergillus conidia may be inhaled continually, providing multiple time points for initiation of disease, thereby neces-sitating the need for frequent sampling but also increasing the possibility of false positivity by detecting Aspergillus airway contamination or colonisation. Although 25% of BAL specimens from healthy volunteers were PCR positive17, the meta-analysis showed that specificity was not significantly affected in a clinical setting14. It is likely that PCR positivity in BAL specimens taken from areas of radiologically confirmed infection will have greater clinical significance because of higher fungal burdens16. So, to confirm a diagnosis of IA at a specific time point, the testing of computed tomography directed BAL specimens may provide the earliest opportunity for genus or species level detection.
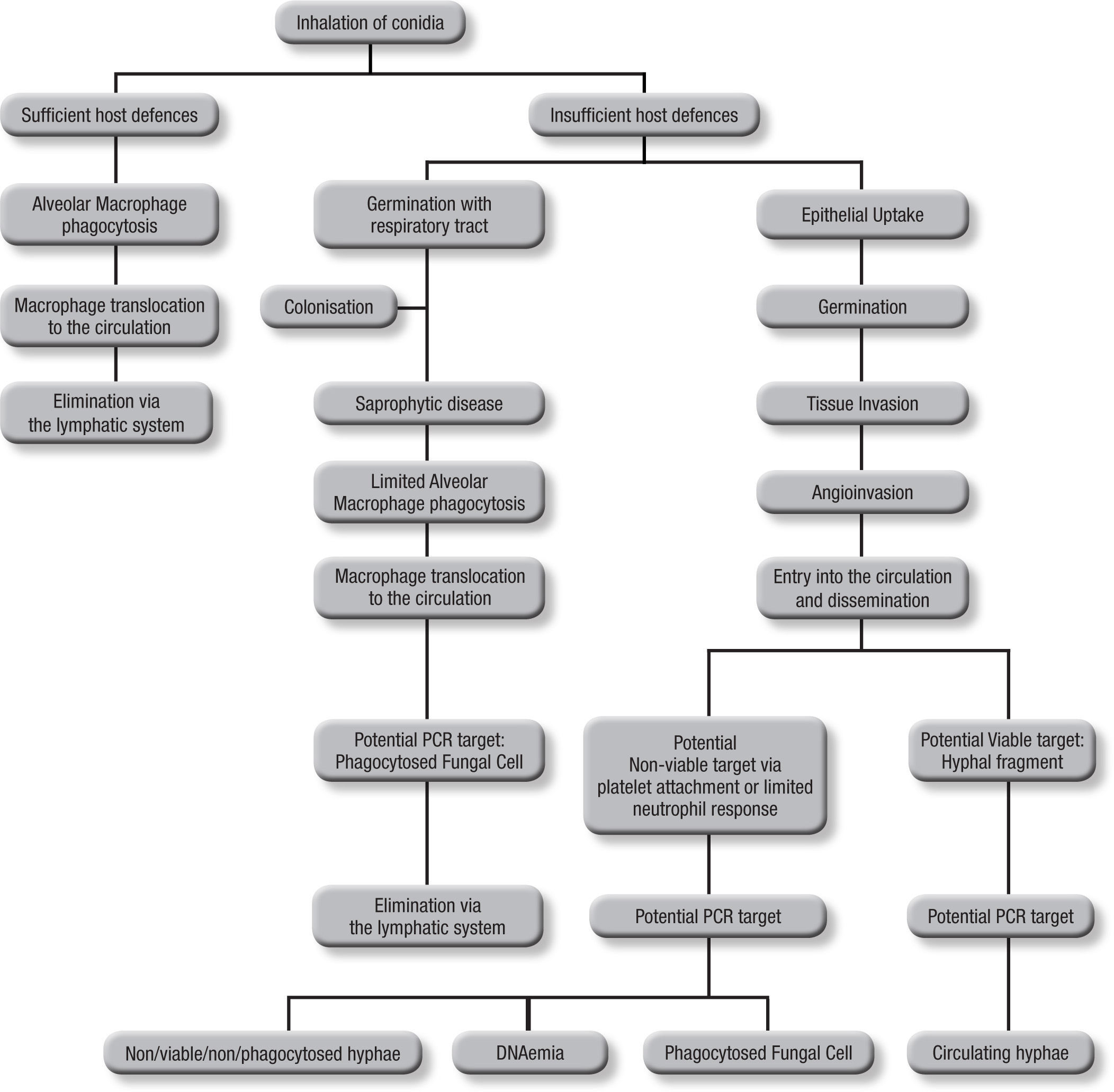
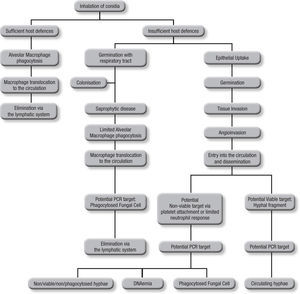
Blood samplesAs the incidence of IA is relative low, (<10%) even among those cohorts considered to be at high risk, it can be argued that infrequent testing of BAL samples is not an optimal strategy. With a low pre-test probability of disease, assays are better suited to excluding disease, relying on a high sensitivity and negative predictive value, in other words “screening”. This approach also reduces the need for unnecessary empirical therapy, thus reducing both cost and antifungal associated patient toxicity18. Taking isolated samples can result in low positivity rates, and this is compounded by the low fungal burdens in certain sample types. Even when a sample is positive, a second sample is needed for confirma-tion to improve the positive predictive value of PCR5. Blood is readily obtained but it is important to consider the potential targets and their route of entry in to the circulatory system (Fig. 1). Normally, alveolar macrophages would clear inhaled conidia, but failure to do so would result in germination and hyphal production that would have been countered by the actions of the neutrophil, but these cells are absent during neutropenia19. Infection will ensue through tissue invasion and angioinvasion, and may coincide with the release of fungal elements such as GM and β-D-glucan and possibly even DNA. If unchecked, infection can then proceed to disease with tissue destruction, haemorrhage and vascular infarction providing the typical early radiological nodule or halo signs. Dissemination may also ensue. This simplistic description of disease would indicate that any fungal target in blood would be dependent on angioinvasion, with potential positivity being dependent on burden, itself being reliant on the degree of angioinvasion. It is also possible that conidia phagocytosed within macrophages could be detected in the circulation shortly after exposure to the organism, which is suggested by the detection of inhaled labelled micro-particles in the bloodstream20. Detecting this potential source could help pre-empt disease, although false positivity could occur, as not every high-risk patient exposed to Aspergillus conidia develops IA. Nevertheless, knowing that a patient at high-risk of developing IA has become infected, to a degree, provides clinically useful information, particularly if combined with host factors (prolonged steroid use or neutropenia, allogeneic stem cell transplantation, graft versus host disease, genetic susceptibility to disease).
The actual source of DNA within the circulation is yet to be determined. Blood cultures seldom yield Aspergillus in cases of IA, yet if seeded, Aspergillus will grow in these cultures, indicating a non-viable source in blood21. Opposing this is the fact that the disease disseminates via the circulation, and for this to occur a viable organism must be available. Morton et al. recently showed that Aspergillus would not grow or replicate in whole blood, and it is possible that, unlike candidaemia, where the yeast is actively growing and budding, Aspergillus enters the circulation but remains relatively inactive until it reaches its tissue destination22. Consequently, the circulatory burden of the mould is likely to be well below the detection limits of current blood culture systems. Non-viable targets, including hyphae damaged by platelet attachment23 and fungal cells phagocytosed within leukocytes probably provide a cell associated DNA source, and free circulating DNA (DNAaemia) will be present when released through the actions of the immune system. DNAaemia could also be influenced by antifungal therapy through its actions on the fungal cell wall/membrane, artificially increasing the DNA burden despite an efficient therapeutic response. In effectively treated cases, this effect would be temporary, as decreasing burden and concomitant rapid circulatory clearance of DNA should result in decreasing burdens. These different targets (free and cell associated DNA) would be found in different components of blood, cell associated DNA would be associated with the leukocytes, if present, whereas free DNA would be found in the serum or plasma, requiring different DNA extraction techniques.
Other samplesHaematogenous transfer can lead to disseminated disease infecting organs and other anatomical structures. This is why histological diagnosis remains the gold standard for diagnosis, but the invasive procedures necessary to obtain samples can be difficult during life. Moreover, PCR testing of these samples is limited and these specimens are usually obtained once, only permitting a diagnostic approach. It is of paramount importance to digest the surrounding tissue efficiently when testing tissue specimens in order that invading hyphae can be targeted. Providing QC for these processes is difficult as developing simulated control samples is limited to specimens obtained from animal infection models or previously tested clinical material.
With cerebral disease, PCR testing of cerebrospinal fluid may be productive, and several studies have shown successful applications24,25. Again testing is limited to diagnosis, as studies and specimens are limited. It is unclear what the actual target within the cerebrospinal fluid sample is, organism or free DNA.
In using PCR to diagnose disseminated disease, it can be argued that the clinical utility of PCR testing will be minimal, the disease is advanced and patient prognosis will be worse compared to testing samples capable of detecting the organism at exposure or infection. However, diagnosis still has value when the disease continues to progress despite antifungal therapy, as it might be due to other moulds such as those belonging to the Mucorales. In this scenario Aspergillus PCR testing should remain negative and pan-fungal PCR or PCR specific to the other aetiology would be required to confirm IFD.
The European Aspergillus PCR Initiative strategyAt the start of the EAPCRI it was decided that using PCR for screening provided the optimal strategy which obviously excluded the use of BAL, cerebrospinal fluid or tissue samples, and focussed on testing blood. With the potential targets located either in the cells or in the plasma/serum it was decided to initially focus on whole blood to pursue the goal of EAPCRI as there was a need for greater standardisation of DNA extraction methods. On completing this process, it was repeated for serum, with plasma being reserved for future investigations.
Whole blood standardisation10The next stage was to evaluate methods currently in use. For any molecular procedure there are two basic steps; first, nucleic acid extraction and then amplification. While it is important to evaluate the combined performance of these processes it is also important to determine the performance of the individual processes; this overcomes the potential effect of combining a poor extraction technique with an optimal PCR test, leading to poor PCR performance.
The first EAPCRI panel was developed to do both, and comprised a panel of quantified A. fumigatus genomic DNA to evaluate PCR alone, and a whole blood panel seeded with different concentrations of A. fumigatus conidia to evaluate DNA extraction and PCR10. Although conidia are unlikely to be involved in the invasive disease process, the panel needed to be consistently quantifiable between centres and it was not possible to accurately quantify multinucleate hyphae. To maintain the goal of a screening assay with a high negative predictive value, the panel was designed to determine limits of detection and overall sensitivity, rather than specificity, and a designated threshold for detection was set. Participants were asked to return results and also supply technical infor-mation for both nucleic acid extraction and PCR amplification to allow subsequent analysis. All data and statistical analyses were performed in a blinded fashion to anonymise the identity of the participant and allow unbiased evaluation.
The DNA panel showed that 90% of participating centres attained the designated threshold. However, when DNA extraction was combined with PCR only nine (41%) of 22 centres reached the same level of performance, indicating that DNA extraction was the rate-limiting step. Enhanced performance was noted with centres processing the entire 3mL sample and the use of “bead-beating” to lyse fungal cells; these recommendations were provided to every recipient of the second panel. This time Aspergillus PCR performance was significantly improved compared to the first round, and performance for centres compliant with the recommendations provided showed further improvements, with a sensitivity and specificity of 88.7% and 91.6%, respectively. Metaregression analysis showed positive associations between sensitivity and compliant methods, the use of white cell lysis, “bead-beating”, and the incorporation of an internal control PCR. A negative association between sensitivity and the use of elution volumes >100μL was also noted. Theseformed the basis of the recommendations for Aspergillus PCR testing of whole blood samples (Table 1.)
European Aspergillus PCR Initiative recommendations for PCR testing of whole blood. Adapted from J Clin Microbiol.2010;48:1231–40.10
| Recommendation | Evidence base |
| Only use EDTA-anticoagulated whole blood specimens | Heparin has been associated with PCR inhibition31 and sodium citrate has been shown to be contamination with Aspergillus32 |
| Please note: batches of EDTA blood collection tubes have been contaminated with Aspergillus DNA and screening of all reagents, including vacutainers, should be performed prior to clinical use33 | |
| Aspergillus DNA and screening of all reagents, including vacutainers, should be performed prior to clinical use33 | |
| Use a minimum of 3mL sample | Two thirds of centres attaining the designated threshold in the EAPCRI studies used 3mL of sample In an analytical study using lower volumes was associated with impaired reproducibility of detection34 |
| Red cell lysis improves performance | >Two thirds of centres attaining the designated threshold in the EAPCRI studies used red cell lysis. In an analytical study not using red cell lysis resulted in a significant delay in Cq values34 |
| The use of white cell lysis is critical | A significant positive association between sensitivity and white cell lysis was determined (P:0.018) |
| Fungal cells should be lysed using bead-beating | A significant positive association between sensitivity and bead-beating was determined (P:0.033). It is also cheaper and quicker than an enzymatic approach and as has been shown to effective on hyphae and conidia35 |
| Screen all reagents before using for routine clinical diagnosis | Contamination of various reagents used in molecular biology with fungal DNA has been noted33,36,37 |
| Use positive and negative extraction controls | Good molecular practice to monitor for inter-assay variation |
| Elute DNAin <100μL | A significant negative association between sensitivity and elution volumes >100μL was determined (P:0.035) |
| PCR testing should be performed in duplicate | Although most PCR assays perform to a similar standard, the fungal burden in clinical samples is likely to be at levels close to or beyond the reproducible limit of detection, consequently replicate testing may be necessary to avoid the reporting of false negative results |
| An internal control PCR should be performed | Good molecular practice to monitor for PCR inhibition and avoid reporting false negative results |
| The internal control should generate a Cq value typical to values seen when testing Aspergillus positive clinical samples | If the burden of internal control target is much higher than the target the affects of inhibition may be less evident, potentially resulting in only slight Cq delays of the internal control. As the fungal burden in blood is at the limit of QPCR detection any delays may result in false negative results |
| Human DNA should not be used for an internal control target | The amount of human DNA will vary per specimen, consequently PCR amplification is purely qualitative and it is not possible to have a reference Cq value from which inhibition can be determined. The burden of human DNA is likely to be far greater than the Aspergillus DNA and may not be as affected by inhibition (See above) |
DNA: deoxyribonucleic acid; EAPCRI: European Aspergillus PCR Initiative; EDTA: ethylenediaminetetraacetic acid; QPCR: quantitative polymerase chain reaction.
On completing whole blood PCR standardisation it was decided to evaluate serum samples. Serum PCR, theoretically, requires less standardisation as there are no human blood cells, fungal cell lysis is no longer required, thus minimising any additional processing steps, and the potential target was likely to be free circulating DNA (DNAaemia), yet the sample still allows a screening strategy to be employed11. It was hypothesised that when targeting such a source, simple nucleic acid extraction protocols could be used, with commercial kits providing greater standardisation and QC; this would be a necessary prerequisite if PCR was to enjoy widespread use outside specialist molecular mycol-ogy laboratories. Following the algorithm described above, a panel of serum samples loaded with varying amounts of A. fumigatus genomic DNA was distributed to laboratory working group members to evaluate protocols currently being used. Again the panel was designed to principally determine analytical sensitivity, both technical information and results were returned, and data analysis was performed blinded to the identity of the participant. As the previous study had shown DNA extraction to be the rate limiting state, a panel to evaluate PCR alone was not included.
Initially, when evaluating whole blood, less than half of methods were able to attain the designated threshold. For serum testing, this figure rose to 82.8% (24/29), confirming the hypothesis that methods testing serum required less standardisation. Overall sensitivity (86.1%) and specificity (93.6%) values were comparable to those for whole blood PCR compliant with EAPCRI recommendations. ROC analysis showed excellent performance, with area under the curve of 0.915, and optimal performance using a Cq threshold of 43 cycles, in keeping with a previous report26. Therefore, it was deemed unnecessary to evaluate procedural recommendations. Instead, meta-regression analysis was performed using the existing data. Significant positive associations were noted between sensitivity and the use of larger sample volumes (>0.5mL) (P=.023), an internal control PCR (P=.029) and PCR assays targeting the ITS region (P=.013). Significant negative associations were noted between sensitivity and eluting nucleic acid in volumes >100 μL (P=.003) and using PCR assays targeting mitochondrial regions (P=.010). However, associations between sensitivity and specific genomic regions, whether positive or negative, should be interpreted with caution as the number of assays targeting the regions stated were limited and reaction kinetics will vary according to the assay design and optimisation, and performance may not be directly associated with the target gene but exclusive to the individual assay11. These results were the basis for the EAPCRI recommendations for PCR testing of serum (Table 2).
European Aspergillus PCR Initiative recommendations for PCR testing of serum.
| Recommendation | Evidence base |
| Use a minimum of 0.5mL sample | A significant positive association between sensitivity and sample volume was determined (P:0.023) |
| Most commercially available nucleic acid extraction systems can be used to extract Aspergillus DNA using the protocols as described by the manufacturer | Twenty-eight of 29 protocols evaluated were able to reach the designated threshold of detection on at least one occasion, generating a threshold positivity rate of 91.5% |
| Screen all reagents, including commercially sourced kits and reagents, before using for routine clinical diagnosis | Contamination of various reagents used in molecular biology with fungal DNA has been noted33,36,37 |
| Use methods specific for testing serum | Methods specifically designed to test whole blood were not able to efficiently process serum samples |
| Use positive and negative extraction controls | Good molecular practice to monitor for inter-assay variation |
| Elute DNA in <100μL | A significant negative association between sensitivity and elution volumes >100μL was determined (P:0.003) |
| PCR testing should be performed in duplicate | Although most PCR assays perform to a similar standard, the fungal burden in clinical samples is likely to be at levels close to or beyond the reproducible limit of detection, consequently replicate testing may be necessary to avoid the reporting of false negative results |
| An internal control PCR should be performed, and for serum testing can be included at the start of the process | Good molecular practice to monitor for PCR inhibition and avoid reporting false negative results. By incorporating the internal control at the start of the extraction process the efficiency of extraction for individual samples can also be monitored |
| The internal control should generate a Cq value typical to values seen when testing Aspergillus positive clinical samples | If the burden of internal control target is much higher than the target the affects of inhibition may be less evident, potentially resulting in only slight Cq delays of the internal control. As the fungal burden in blood is at the limit of QPCR detection any delays may result in false negative results |
| Human DNA should not be used for an internal control target inhibition can be determined | The amount of human DNA will vary per specimen, consequently PCR amplification is purely qualitative and it is not possible to have a reference Cq value from which |
| Use a PCR positivity threshold of 43 cycles | This provides the greatest degree of diagnostic accuracy (DOR of 105) |
DNA: deoxyribonucleic acid; QPCR: quantitative polymerase chain reaction.
The obvious next step for the EAPCRI would be to test plasma, as this is the complete liquid component of blood without the cell, and includes the clotting factors. Theoretically, both serum and plasma types contain the same target (DNAaemia), but the blood clot formed prior to obtaining serum could, in addition to trapping human erythrocytes, leukocytes, and any fungal cells potentially reduce the amount of free DNA in the sample; this would not happen in plasma. Evaluating the effect of these different sample types on the mechanics of any nucleic acid extraction tech-nique would be relatively straightforward through testing plasma and serum samples loaded with the same fungal DNA burdens after the separation of the required blood fractions. However, this would not assess the affect of clot formation on the DNA burden. This could be assessed using simulated samples, spiking blood collection tubes with Aspergillus DNA, possibly prior to drawing blood, but certainly prior to clot formation, and for plasma testing by spiking the ethylenedi-aminetetraacetic acid blood tube prior to fractionation.
For confirming the diagnosis, specimens require more invasive procedures, and obtaining these samples must be weighed up against the potential benefit to the patient, and whether a PCR diagnosis will alter patient management or outcome. For instance, if a patient is already receiving antifungal therapy and is responding, then obtaining a deep tissue sample for PCR testing may not lead to a change in therapy or alter the outcome. Post-mortem testing could confirm a diagnosis, but there is no need for rapid diagnosis as a histopathological/mycological approach will suffice. The enhanced sensitivity and specificity provided by realtime PCR provides the opportunity to pre-empt infection from becoming manifest disease, thereby helping improve the patient prognosis. Currently, since most cases of IA begin in the lung, obtaining a BAL specimen is the only way of attaining an appropriate specimen for diagnosis. Establishing testing panels with this specimen presents several logistic hurdles, including sourcing “willing” volunteers to provide sufficient BAL samples, and overcoming potential airway contamination leading to Aspergillus positive BAL material. The potential of evaluating a simulated BAL specimen is currently being pursued.
The EAPCRI also recognises the contributions of other organisations trying to improve the quality of molecular diagnosis and lead to greater standardisation. The Invasive Aspergillosis Animal model group are developing standardised in vivo models of IA to provide representative pathological samples, while the Aspergillus Technology Consortium is developing a bank of samples from patients with IA to provide source material for evaluating diagnostic tests.
Commercial assays are now becoming available, and preliminary clinical validation looks promising, although patient numbers are limited when compared to many evaluations of “in-house” protocols27,28. From the beginning the EAPCRI has sought commercial collaboration and by participation in testing EAPCRI QC panels, commercial partners ensure their analytical performance is comparable to well validated methods. The commercial production and the independent analytical performance validation of commercial assays provide the opportunity for Aspergillus PCR to be performed outside specialist molecular mycology laboratories.
The minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines were published as a guide to researchers as to the minimum information required in manuscripts describing real-time PCR experiments29, and this has recently been applied specifically to an Aspergillus PCR protocol30. While we fully support this guidance, it is important to add that the MIQE guidelines were published to encourage researchers to provide the technical information necessary for readers to compare and replicate assays. It is unwise to assume that these recommendations have been followed in papers published before these guidelines appeared however, limitations in space often precludes the inclusion of extensive technical information, even if assay optimisation and analytical evaluation has been performed as described by the current MIQE guidelines. There needs to be direct comparisons of the recent MIQE designed Aspergillus PCR assay with other previously well validated assays before concluding that it will provide improved clinical performance, particularly when our research shows DNA extraction to be rate-limiting10.
In conclusion, the EAPCRI has made significant steps in developing a standard for Aspergillus PCR, but recognises that the process will not be finished until the clinical utility has been established in formal clinical trials. Currently, samples for PCR testing are being taken in two prospective trials, one comparing pre-emptive and empirical strategies (EORTC 65091–06093: ClinicalTrials.gov Identifier: NCT01288378), and the other on the prevention of invasive fungal infections in subjects receiving chemotherapy for acute lymphoblastic leukaemia (Ambiguard: ClinicalTrials.gov Identifier: NCT01259713). Methodological recommendations are being evaluated using non-simulated animal model material, and collaborations with Invasive Aspergillosis Animal model group continue. On completion, it is hoped that PCR will have attained sufficient standardisation and validation to be included in the EORTC/ MSG definitions of IFD.
Conflicts of interestAll authors declared no conflicts of interest.