Coagulase-negative staphylococci (CoNS) are among the main causative agents of bacteremia. Staphylococcus epidermidis and Staphylococcus haemolyticus are the CoNS species most frequently isolated. These species are often associated with infections in immunocompromised patients who have a medical device implant. Methicillin resistance was first described in Staphylococcus aureus; it has also been reported in CoNS species. Methicillin resistance is conferred by expression of the mecA gene, contained within the staphylococcal cassette chromosome (SCCmec). mecA gene codes for the PBP2a protein which shows poor binding to beta-lactam antibiotics, so methicillin-resistant strains are resistant to beta-lactams. The presence of SCCmec in CoNS species complicates infections caused by these organisms, since it confers resistance to a variety of antibiotics, making treatment difficult. This review analyzes the clinical relevance of SCCmec as well as the diversity and structure of elements present in CoNS species.
Coagulase-negative staphylococci (CoNS) are key agents contributing to infections in hospital environments. They are microorganisms which form part of the normal skin and mucus membranes of humans, frequently isolated from patients with skin alterations, for instance, subjects undergoing surgery or with trauma injuries. Even though they are sometimes considered as culture pollutants in clinical bacteriology laboratories, they are currently considered opportunist pathogens. They have also been isolated from patients with implanted devices, such as catheters, prosthesis, derivations, etc.1–3
In our hospital, studies on the frequency of isolated microorganisms in medical device-related infections have been conducted before. An example of which is a study of the incidence of infections related to central venous catheters (CVC) in the intensive care unit (ICU), where a group of CoNS was the second most frequent group (8.8%) in this type of infection, only exceeded by Staphylococcus aureus.4 Similarly, in a study of infectious endocarditis cases, CoNS were the most frequent causative agent after S. aureus.5
In diverse studies, CoNS have been classified as the main causative agent of bacteremia, causing around 30% of bacteremia cases confirmed by the laboratory.6 Within this group, Staphylococcus epidermidis and Staphylococcus haemolyticus are two CoNS species most frequently isolated in bacteriology laboratories.7 These agents are the most frequent cause of bacteremia. Also, these species present high percentages of resistance to methicillin, a characteristic which converts infections produced by these species into difficult-to-treat pathologies.8 In this revision, we will discuss the clinical relevance of CoNS, as well as the genetic elements responsible for methicillin resistance of CoNS, particularly S. epidermidis and S. haemolyticus.
Resistance to methicillinOriginally, penicillin was the pharmaceutical of choice for the treatment of infections caused by S. aureus. Soon after, penicillin-resistant strains emerged due to the acquisition of plasmids which codify β-lactamases or penicillinases.
Methicillin was introduced in 1960: a semisynthetic penicillin resistant to penicillinases. Nevertheless, a year later the first methicillin-resistant strains appeared. Consequently, the term methicillin-resistant S. aureus was coined (MRSA). MRSA strains present an additional resistance to the other β-lactamases antibiotics, including cephalosporins and cephemic agents.9
Resistance to methicillin has also been reported in CoNS species isolated from clinical samples, with proportions even higher than those reported in S. aureus. There are reports of methicillin-resistant S. epidermidis strains (MRSE) and methicillin-resistant S. haemolyticus (MRSH), isolated from blood, with resistance percentages over 70% for both species.10–12 Also, MRSE and MRSH have been isolated from healthy subjects, who are colonized, thus constituting an important dissemination source for these species.13,14
Mechanism of resistance to β-lactams antibiotics in methicillin-resistant strainsPenicillin binding proteins (PBP) are proteins involved in the metabolism of peptidoglycan of the bacterial cell wall.15 When β-lactam antibiotics enter the active PBP site they are bound covalently, inhibiting PBP function, hence the wall synthesis.16
Methicillin-resistant strains produce a penicillin-binding protein, known as PBP2a or PBP2′ which presents a low affinity to β-lactams.16,17 Therefore, antibiotics do not remain attached to PBP2a, inhibition of the peptidoglycan synthesis does not occur, and cellular lysis is avoided.16
PBP2a protein is coded by the mecA gene, which is contained in a mobile genetic element called staphylococcal cassette chromosome mec (SCCmec).18 SCCmec was first described in an MRSA strain, whose sequencing set the bases for the study of SCCmec basic characteristics, as well as its diversity.
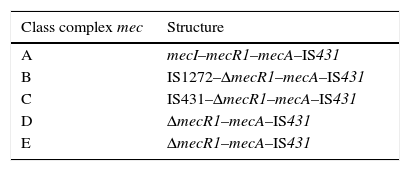
SCCmec characteristicsSCCmec is a mobile genetic element which attaches in the bacterial chromosomal attachment site (attBSC), near the origin of replication in staphylococci. Moreover, SCCmec possesses insertion sequences (IS) which flank the mecA gene, as well as expression-regulator genes. The set of regulator regions, insertion sequences and mecA gene, are called a mec complex. Several types of mec complexes have been described, depending on the IS type present in the mecA environment. So far, five types of mec for S. aureus (complex A to E) have been described.19mec complex is formed by mecA, regulator genes mecI and mecR1, as well as different insertion sequences which may partially delete regulator genes.
Class A mec complex is the prototype complex (Table 1); it contains the gene mecA and gene regulators mecI and mecR1. In class B mec complex, the gene mecR1 is partially deleted by the insertion of IS1272; while in class C mec complex the gene mecR1 is partially deleted by the insertion of IS431. Class C mec complex is divided into two variants, C1 and C2, depending on the orientation of IS431. Class D mec complex presents the gene mecR1 deleted, but without an associated insertion sequence. Class E mec complex is similar to the latter, but with a greater rate of mecR1 deletion.19,20 All complexes present an IS431 subsequent to mecA.
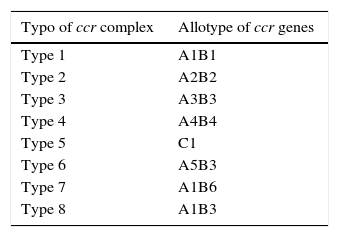
Also, SCCmec possesses genes which codify for recombinases. These recombinases are in charge of integrating the SCCmec in the bacterial chromosome. The recombinase gene groups are called ccr. There have been three different types of recombinases to date: ccrA, ccrB, and ccrC; coded by the genes ccrA, ccrB and ccrC, respectively. There are different allotypes for each one, depending on the existent homology degree.21 Various allotypes have been identified for recombinase A and B, while just one allotype has been described for recombinase C.20 They have been classified into eight different types of complex ccr depending on existing recombinases and allotypes in the different cassettes (Table 2).
The regions located between the complex ccr, complex mec and SCCmec ends are known as J regions (joining regions), containing additional structures such as plasmids or IS, which have the ability to provide additional resistance characteristics.20 The J1 region is located between the right chromosomic binding and the ccr complex. The J2 region is located between the ccr complex and the mec complex. The J3 region is located between the right chromosomic binding and the mec complex.
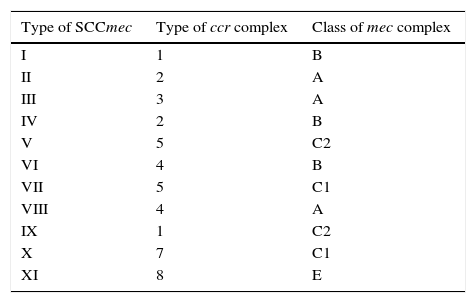
SCCmecs are classified into types and subtypes. SCCmec types are defined by the type of mec complex and ccr complex present. Subtypes are based on differences present in J regions (joining regions) within a SCCmec. These differences may be plasmids, insertion sequences, or pseudogenes.19 Thus far, eleven types of SCCmec in S. aureus have been described, based on the combination in mec complex class and ccr complex type (Table 3).21–25
SCCmec and resistance to antibioticsDifferent genetic elements, such as genomic islands, bacteriophages, pathogenicity islands, chromosomic cassettes, plasmids, insertion sequences and transposons, intervene in the dissemination of resistance genes and staphylococci virulence.
Insertion sequences are particularly capable of carrying resistance determinants through homologous recombination. CoNS-resistant strains can function as a deposit for antibiotic-resistant genes, which may be transferred to microorganisms, including the S. aureus strains.26
With the exception of mecA, type I, IV, V, VI and VII, SCCmec do not contain antibiotic-resistant genes. However, type II SCCmec bears the Tn554 transposon, which contains erythromycin-resistant genes (ermA) and spectinomycin (spc). This SCCmec also contains plasmid pUB110 (flanked by a pair of IS431 elements) which codifies resistance to kanamycin, tobramycin and bleomycin.
Similarly, type III SCCmec contains transposon Tn554, plasmid pUB110 (with the resistance determinants previously mentioned), an integrated copy of pT181 (confers resistance to tetracycline) and pI258 (resistance to mercury).27 Type IX and X SCCmec contain determinants resistant to cadmium, copper and arsenate. Type XI SCCmec bears resistance to arsenate.
SCCmec in CoNSIn addition to the types and subtypes of SCCmec, there are variations of SCCmec and non-typeable SCCmec observed in several studies, mainly in CoNS, which means that CoNS have a wide reservoir of SCCmec structures. In the case of an SCCmec with a non-typeable ccr complex, there have been reports of mec complexes existing even in the absence of the known types of ccr.28,29 Moreover, the existence of new versions of recombinases has been suggested, hence it is not possible to detect them with the methodology established for S. aureus.
mec and ccr complexes go through complex recombining and reordering processes in CoNS genomes, through which the generation of new SCCmec types occurs. Most likely just a small fraction of the SCCmec diversity is present in S. aureus.
SCCmec IV is the most dominant in MRSA infections acquired in the community (CA-MRSA).30 However, SCCmec IV is also frequently found in CoNS, particularly in S. epidermidis, in studies of the community and hospital environment. In a report on hospital isolations of S. epidermidis, 36% was type IV SCCmec, 34% type II SCCmec, 28% type III SCCmec and just 2% type I SCCmec.31 Another study conducted in healthy patients found a frequency of 57% of type IV SCCmec.14 Additionally, other types of SCCmec in S. epidermidis have often been reported. Such is the case of type III SCCmec which has been reported in clinical and community isolations.31,32
Some studies suggest the transference of SCCmec between S. epidermidis and S. aureus.31 Type IV SCCmec in S. epidermidis shows a homology of 98–99% with type IV SCCmec in S. aureus. In addition, resistance to methicillin is highly frequent in S. epidermidis isolations and less frequent amongst S. aureus isolations (over 70% of S. epidermidis hospital isolations are resistant to methicillin). Hence, it is possible for SCCmec diversity to be greater in S. epidermidis.31,33 In a study conducted by Wisplinghoff et al., type IV SCCmec was highly frequent amongst S. epidermidis isolations during the seventies and was not found among MRSA isolations recovered during this time. However, the first S. aureus with type IV SCCmec was recovered during the eighties.31 Type V SCCmec is also distributed among CoNS.34 The characterization of S. haemolyticus SCCmec has proven that type V SCCmec is the most frequent amongst this species, even though there have also been reports of other SCCmec structures.33,34
Nosocomial frequency of methicillin-resistant S. haemolyticus may be as high as 90%35, while also being resistant to most antibiotics. As for S. epidermidis, from the association with type V SCCmec emerges the hypothesis of transference from said SCCmec type to S. aureus, which has been shown through SCCmec sequencing in both species, resulting in a homology superior to 99%, thus making the transference between species possible, which would involve S. haemolyticus as a reservoir of type V SCCmec.35
The loss of ccr complex has been demonstrated in S. haemolyticus, which may contribute to the resistance phenotype providing S. haemolyticus the ability to retain the resistance to methicillin and resistance to other antimicrobial agents contained in SCCmec. Additionally, over 82 insertion sequences have been detected in S. haemolyticus, which are thought to provide a high genomic plasticity to the species.29
It has been suggested that class C mec complex and ccrC complex were originated in S. haemolyticus; and that type V SCCmec and other SCCmec variants were generated in this specie. This hypothesis is frequently supported by the finding of 99% of nucleotide identity amongst class C mec complexes identified in MRSA and SH-MR isolations.29,35
Barros et al34 reported 43% (24 isolations) of S. haemolyticus isolations as non-typeables. Therefore, it is clear that the diversity of elements in the S. haemolyticus genome is really large, thus the best way to know this diversity would be through their sequencing.
ConclusionsResistance to methicillin (with the subsequent resistance to a variety of pharmaceuticals) is a worldwide distribution problem which occurs in community-related infections as well as in nosocomial infections. The increase in the frequency of infections caused by multi-resistant microorganisms makes the study of distribution and diversity of genetic elements which confer resistance to pharmaceuticals in bacteria of clinical importance necessary, as well as the study of transmission mechanisms of such elements.
Conflict of interestThe authors have no conflicts of interest to declare.