Multiple sclerosis (MS) is a demyelinating disease that affects young adults; in that age group, it represents the second leading cause of disability in our setting. Its precise aetiology has not been elucidated, but it is widely accepted to occur in genetically predisposed patients who are exposed to certain environmental factors. The discovery of the regulatory role played by intestinal microbiota in various autoimmune diseases has opened a new line of research in this field, which is discussed in this review.
DevelopmentWe reviewed published studies on the role of the microbiota in the development of both MS and its animal model, experimental autoimmune encephalomyelitis (EAE). In mice, it has been shown that intestinal microorganisms regulate the polarisation of T helper cells from Th1-Th17 up to Th2, the function of regulatory T cells, and the activity of B cells; they participate in the pathogenesis of EAE and contribute to its prevention and treatment. In contrast, evidence in humans is still scarce and mainly based on case-control studies that point to the presence of differences in certain bacterial communities.
ConclusionsMultiple evidence points to the role of microbiota in EAE. Extrapolation of these results to MS is still in the early stages of research, and studies are needed to define which bacterial populations are associated with MS, the role they play in pathogenesis, and the therapeutic possibilities this knowledge offers us.
La esclerosis múltiple (EM) es una enfermedad desmielinizante que afecta a adultos jóvenes, grupo en que supone la segunda causa de discapacidad en nuestro medio. Su etiología precisa no está dilucidada, pero se acepta que se presenta en pacientes predispuestos genéticamente que se ven expuestos a determinados factores ambientales. El descubrimiento del papel regulador de la microbiota intestinal en diversas enfermedades autoinmunes ha abierto una nueva línea de investigación en este campo, lo que se discute en esta revisión.
DesarrolloRevisamos los estudios publicados acerca del papel de la microbiota en el desarrollo de la EM y su modelo animal, la encefalomielitis autoinmune experimental (EAE). En ratones, se ha demostrado que los microorganismos intestinales regulan la polarización de las células T helper de Th1-Th17 hasta Th2, la función de las células T reguladoras y la actividad de las células B, participando en la génesis de la EAE, así como en su prevención y tratamiento. Por el contrario, en humanos la evidencia es aún escasa, fundamentalmente en base a estudios de casos control que apuntan a la existencia de diferencias en determinadas comunidades bacterianas.
ConclusionesExiste múltiple evidencia del papel de la microbiota en la EAE. La extrapolación de los resultados a la EM está en las primeras fases de investigación, y hacen falta estudios que definan qué poblaciones bacterianas se asocian a la EM, su papel en la patogenia y las posibilidades terapéuticas que esto nos ofrezca.
Multiple sclerosis (MS) is a demyelinating disease characterised by inflammatory lesions at different locations in CNS white matter resulting in glial scars, demyelination, oligodendrocyte loss, and axonal damage.1 MS affects over 2.5 million people worldwide and around 46000 people in Spain, and it has an estimated prevalence of approximately 100 cases per 100000 population.2 As with other autoimmune diseases, the incidence of MS has increased in recent years.3 MS usually appears in young adults4 and represents the second most frequent cause of disability in this population in our setting (after trauma).5 The age of onset, combined with a long life expectancy (6 years less than the general population),6 results in long disease progression times frequently exceeding 30 years,7 which has considerable clinical, social, and economic consequences.
Although the exact pathophysiology of MS is yet to be fully understood, epidemiological data indicate that genetic and environmental factors play an important role.8,9 Thus, MS may present in genetically predisposed individuals who are exposed to certain environmental factors,10 especially during childhood.11 Among these are such well-studied factors as Epstein–Barr virus infection,12–15 smoking,16–18 latitude,8 and vitamin D levels.19–21 Recent studies are evaluating other less known factors, including sodium intake,22,23 BMI during adolescence,12 levels of leptin (hormone produced by adipose tissue),24 vitamin A levels,25 alcohol use,26 and other factors related to the “hygiene hypothesis”, for example helminth 27 or Helicobacter pylori infection.28 Other environmental factors being explored are the microorganisms constituting the intestinal microbiota, which we address in this study.
Gastrointestinal microbiotaThe gastrointestinal tract contains over 100 trillion microbes (1014); most are located in the colon (1011-1012) and they outnumber human cells by a factor of 10.2–4 This complex microbial community, known as the gastrointestinal microbiota, consists of bacteria, archaea, eukaryotes, fungi, and viruses.29,30 Some regard humans as “meta-organisms”, since they contain 10 to 100 times more bacterial cells than human cells, and these are metabolically and immunologically integrated.31 Microbiota composition depends not only on location but also on such other factors as age, sex, race, diet, medication (especially antibiotics), stress, smoking, gastrointestinal infections, and individual factors.30–32 Likewise, individuals may exhibit marked temporal variations in microbiota composition.33 Although we currently lack a definition for healthy microbiota, we do know that richness and diversity are indicators of health34 whereas lower bacterial richness is associated with obesity and markers of metabolic disorders.35 Regarding microbiota composition, numerous studies have attempted to correlate certain types of microorganisms to different physiological states. Some studies suggest that certain microorganisms improve metabolism; resistance to infections, cancer, autoimmunity, or inflammation; endocrine signalling; and brain function (gut-brain axis). The following bacteria are considered to be associated with a good state of health: Bacteroides, Bifidobacterium, Clostridium clusters XIVa and IVa (butyrate producers), Eubacterium, Faecalibacterium, Lactobacillus, and Roseburia.32
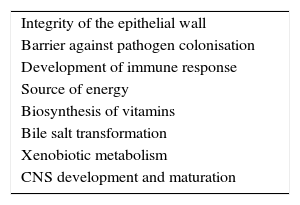
In summary, our understanding of the relationship between microbiota and the human host is redefining itself from the traditional concept of commensalism to the more accurate idea of mutualism: bacteria perform functions in our organism, ranging from metabolic activity to immune homeostasis, which are not genetically coded (Table 1).36,37 The human microbiota may be considered a virtual organ that is essential for immune response. Numerous studies now focus on analysing the role of the intestinal flora in human health, infections, inflammatory diseases, neoplastic disorders, and – as we will address in this study – in autoimmune diseases.38
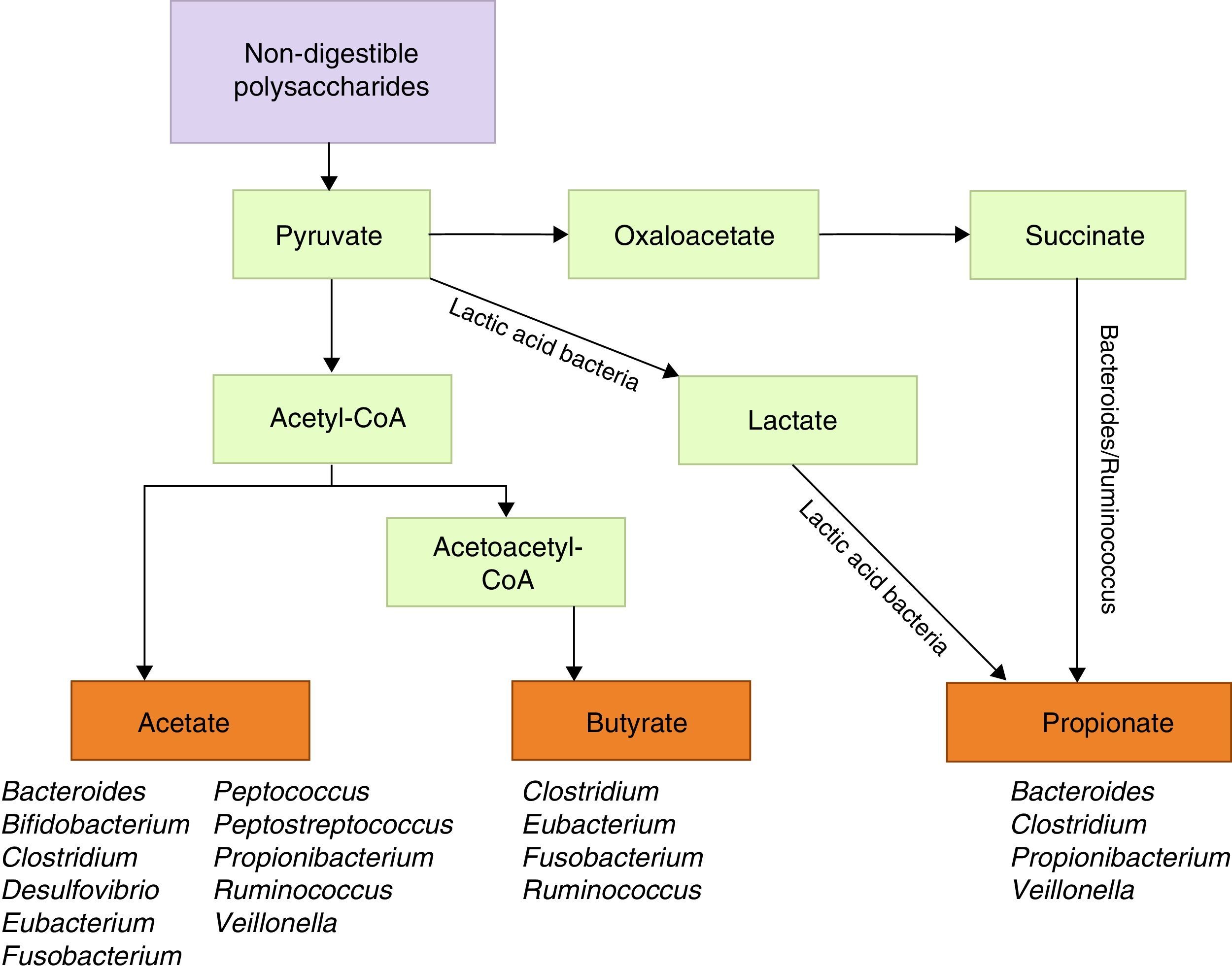
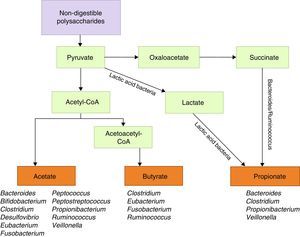
Metabolic functions include degradation of plant polysaccharides into such short-chain fatty acids as butyrate, propionate, and acetate. Since these fatty acids have anti-inflammatory properties and serve as the main energy source for colonocytes, they fortify the intestinal mucosal barrier (Fig. 1).39 Some bacteria within the flora, specifically lactobacilli, contribute to cholesterol metabolism40,41 and the production of vitamin K and different B vitamins.41 They are also active in the metabolism of xenobiotic compounds, drugs, antibiotics, or bioactive products,42 since they affect the pharmacokinetic properties of these compounds; intestinal microbiota also affects the production of certain toxins involved in some types of neoplasia.43,44
The recent concept of “gut-brain axis” refers to the afferent and efferent connection between the brain and the gastrointestinal system by means of nutrients and neuroendocrine and immunological signals.45 The microbiota has been shown to affect CNS function both in healthy and diseased individuals.46 It is involved in brain development47; the hypothalamic-pituitary-adrenal axis48,49; expression of serotonin 5-HT1 receptors; exchange of such neurotransmitters as serotonin, dopamine, and norepinephrine; and changes in proteins regulating neuronal synapse development and function.47 Considering the above, the microbiota is understood to play a role in motor control, anxiety behaviour, and cognitive functions linked to brain development during the early stages of life.50,51
Microbiota and immune responseThe gut microbiota contributes to a variety of immunological functions. In the intestinal barrier, it prevents colonisation and growth of pathogenic microorganisms52,53 and promotes immune barrier maturation by stimulating both innate immune response via toll-like receptors (TLR) and NOD-like receptors54–56 and adaptive immune response; the microbiota also plays a major role in the secretion of mucins, antimicrobial peptides, defensins, and IgA.53 Regarding the development of systemic immune response, studies on germ-free mice have shown that gut microbiota is involved in the regulation and maturation of Peyer patches, mesenteric lymph nodes, and germinal centres. It also regulates the number of IgA-producing plasma cells, intestinal gamma delta T cells, and lamina propria or intraepithelial CD4+ T cells, and it is involved in gene expression of TLRs and major histocompatibility complex class II.53
The gut microbiota is also involved in effector T cell development and cytokine production and has a marked influence on Th17 cells and regulatory T (Treg) cells, which are involved in autoimmune response and its regulation. As such, it is involved in autoimmune diseases in general and MS in particular. Germ-free mice (mice born in germ-free conditions and lacking intestinal flora) show lower levels of Th1 and Th17 cells and increased levels of Th2 cells, but this situation is reversed when mice are colonised by normal intestinal flora.57,58 It has been theorised that microbiota induces conversion of steady-state Th17 cells to pathogenic Th17 cells, which produce IFN-γ to elicit intestinal inflammation. This occurs in the presence of a proinflammatory microenvironment promoted by certain cytokines, such as IL-12, IL-23, IL-1β, and TGF-β3.59,60 A single microbe, segmented filamentous bacterium (SFB), has been found to be sufficient to induce autoimmunity-promoting Th17 activity.61
The gut microbiota is essential for Treg cell development and function.62 These cells use IL-10 to regulate the inflammation caused by microbial stimulation.59 Numerous microbes, especially Bacteroides fragilis (more specifically its polysaccharide A [PSA]), have been associated with Treg cell induction63–65; this microbe has been linked to development of IL-10-producing FoxP3+ Treg cells, and to the prevention and resolution of experimental colitis in animal models; this demonstrates the key role of Treg cells in regulating immune tolerance.66,67
Short-chain fatty acids, especially butyrate, exert an anti-inflammatory effect on the immune system. They increase IL-10 and IL-4 secretion,68 inhibit leucocyte adhesion to endothelial cells via VCAM-1 modulation,69 inhibit IFN-γ function and consequently its proinflammatory activity,70 and regulate Treg cell generation71–73 and leucocyte anti-inflammatory function.74,75
Microbiota and demyelinating diseasesNew evidence suggests that, in addition to affecting brain development, microbiota is also linked to certain neurological diseases, especially autoimmune diseases. There is some evidence to suggest that gut microbiota plays a role in the development of autism spectrum disorders,76 Guillain-Barré syndrome,77 and such psychiatric disorders as depression, anxiety, and schizophrenia.78 Regarding demyelinating diseases, numerous studies have analysed the connection between the intestinal microbiota and neuromyelitis optica and especially experimental autoimmune encephalomyelitis (EAE), the animal model for MS. Furthermore, some studies are now trying to extrapolate results in EAE to MS.
Patients with aquaporin-4-IgG-positive neuromyelitis optica show greater antibody response to gastrointestinal antigens than healthy controls, especially in longitudinally extensive myelitis, which suggests that the gut microbiota controls autoimmune inflammation.79 Other studies have also shown the presence of aquaporin-4-specific T-cells in peripheral blood which exhibit Th17 polarisation and display cross-reactivity to a homologous sequence of an epitope within Clostridium perfringens, a bacteria of the commensal flora; this points to a mechanism of molecular mimicry responsible for activating the previously described Th17 response.80
Microbiota and experimental autoimmune encephalomyelitisThe first evidence of an association between intestinal bacteria and peripheral tolerance and EAE prevention or treatment came from studies in which mice received an oral “vaccine” for enterotoxigenic Escherichia coli composed of a live, attenuated Salmonella typhimurium vector-expressing enterotoxigenic E. coli fimbriae, colonisation factor antigen I (CFA/I). Mice receiving this prophylactic intervention experienced mild EAE and recovered completely. Histopathology studies revealed fewer inflammatory infiltrates in spinal cord grey and white matter as well as decreased expression of IFN-γ-secreting T cells and increased secretion of IL-4, IL-10, IL-13,81 and TGF-β.82 This “vaccine” was found to have anti-inflammatory properties; its mechanism involved stimulating FoxP3+ Treg cells.83,84
Subsequent studies focused on analysing the direct involvement of gut microbiota in the development of EAE. Some studies found an association between microbiota alterations secondary to broad-spectrum antibiotics and improvements in EAE progression. Administering a combination of kanamycin, colistin, and vancomycin before EAE induction halted EAE development and was accompanied by decreased production of proinflammatory cytokines (IL-6, IL-17, IFN-γ, and TNF-α) and lower numbers of mesenteric Th17 cells.85 Ochoa Repáraz et al.86 repeated the experiment using ampicillin, vancomycin, neomycin sulfate, and metronidazole (either injected or orally) and included a control group of untreated mice. This study found that mice receiving antibiotics orally experienced less severe EAE and displayed FoxP3+ Treg cell accumulation in peripheral and cervical lymph nodes, in addition to mesenteric lymph nodes, which suggests that Treg cell regulation occurs in other areas apart from the intestine. CD103+ dendritic cells were found to be involved in this mechanism; there were also increased IL-13 levels and decreased IL-17 levels. Furthermore, the protective effect was lost when animals were recolonised with commensal bacteria. These researchers also demonstrated that antibiotics altered B cell levels in EAE mice by inducing a subpopulation of CD5+ B cells which conferred protection against EAE; in vitro, these cells generated IL-10 when stimulated with proteolipid protein.87 Adoptive transfer of CD5+ B cells from mice treated with antibiotics to untreated mice reduced EAE severity considerably.
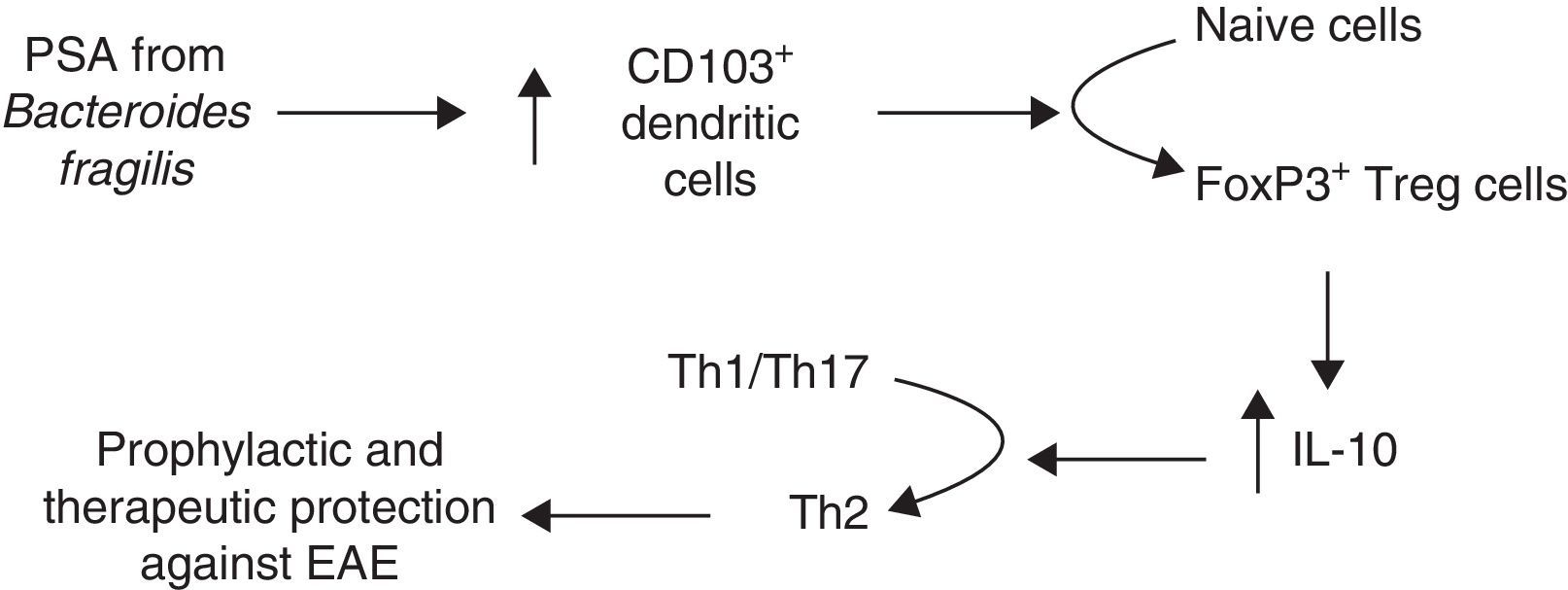
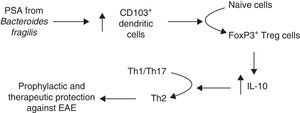
Another line of research analyses the role of B. fragilis PSA, which modulates the immune system by correcting Th1/Th2 imbalances,57 and prevents intestinal inflammatory disease.66 In light of these findings, Ochoa-Repáraz et al.88 tested PSA in EAE. These authors first reconstituted the intestinal flora with either B. fragilis, the wild type able to express PSA, or with PSA-deficient B. fragilis in mice whose flora had been altered with oral antibiotics. Mice recolonised with PSA-producing strains of B. fragilis remained protected against EAE and showed a higher percentage of IL-10–producing FoxP3+ Treg cells. In a subsequent study, Ochoa-Repáraz et al.89 administered purified PSA by the oral route; this antigen was found to have both prophylactic and therapeutic effects against EAE, whereas this effect was not seen in IL-10-deficient mice. Upon observing increased levels of CD103+ dendritic cells and FoxP3+ Treg cells in cervical lymph nodes, these authors proposed a mechanism by which PSA improves disease outcomes in mice (Fig. 2). In 2014 and 2015, these researchers presented 2 studies addressing the mechanism mediating PSA protection against EAE and observed that TLR2 mediates expansion of CD39+ CD4 T cells; likewise, CD39+ confers immune-regulatory phenotypes to T cells in general and FoxP3+ Treg cells in particular.90,91
Other studies have aimed to determine the microorganisms responsible for the disease, for example SFB, which is able to induce Th17 cells.61 Lee et al.92 attempted to induce EAE in germ-free mice, which developed mild EAE and displayed lower levels of IFN-γ and IL-17A in both the intestine and spinal cord, reduced Th1/Th17 proinflammatory responses in the CNS, and high levels of FoxP3+ Treg cells. These authors showed that colonisation with SFB resulted once more in EAE, along with increased IL-17 levels in the intestine and Th17 response in the CNS, colon, and small intestine, which suggests that these bacteria alone may cause neurological inflammation and therefore trigger the disease.
Berer et al.93 used germ-free mice spontaneously developing EAE to demonstrate that both the target autoantigen – myelin oligodendrocyte glycoprotein (MOG) – and microbiota had to present for the disease to develop. Germ-free mice were found to display a decrease in Th17 cells in the intestinal wall, lamina propria, and Peyer patches, as well as decreased production of IFN-γ and IL-17 in the spleen. These authors observed that anti-MOG antibody production decreased in mice with no microbiota and increased rapidly when these mice were recolonised.
Lactic acid bacteria are other common group of commensal bacteria that have been associated with immune response regulation and studied in EAE. This group includes Pediococcus acidilactici. Oral administration of this bacterium induces an IL-10-mediated response leading to decreased EAE severity. It works both therapeutically and prophylactically by inhibiting IL-17 and IFN-γ and decreasing cellular infiltration in the CNS. In this case, T cells involved in this mechanism were Treg1 cells rather than CD4+ FoxP3+ T cells (the latter increased only slightly).94
Recent studies have addressed other non-bacterial microorganisms. A study published in 2015 analysed the impact of the yeast Candida kefyr on EAE and mouse microbiota.95 This yeast led to clinical improvements due to the induction of CD103+ dendritic cells and FoxP3+ Treg cells. We now know that these dendritic cells stimulate Treg cells through retinoic acid and TGF-β.96 The analysis of mouse microflora after administration of C. kefyr revealed increased levels of lactic acid bacteria and Prevotella to the detriment of Bacteroides; the authors attributed improvement to this decrease mediated by reduced IL-6 production.94
Lastly, others have investigated the potential usefulness of probiotics for EAE. Other researchers have administered Bifidobacterium animalis to nursling mice in which EAE was later induced; this resulted in a decrease in symptom duration, curiously enough only in males.97 A combination of 3 Lactobacillus strains, was found to reverse EAE in mice and increase FoxP3+ Treg cells and IL-4, IL-10, and TGF-β1 in mesenteric lymph nodes and the spleen.98 This was not observed for any of the strains used separately. Other probiotic mixtures (Lactobacillus, Bifidobacterium, and Streptococcus) have yielded similar results; this mechanism also involved IL-10 and the development of the Treg cells that inhibit Th1/Th17 polarisation, as mentioned previously.99
In summary, the available evidence (Table 2) strongly supports the association between microbiota and immune response to EAE in light of the gut microbiota's impact on EAE pathogenesis, prevention, and treatment in animal models.100 The immunological mechanisms involved in the regulation of immune response are based on 3 main concepts: Th cell polarisation towards Th1, Th2, or Th17 subtypes; Treg cell function; and B cell activity.101
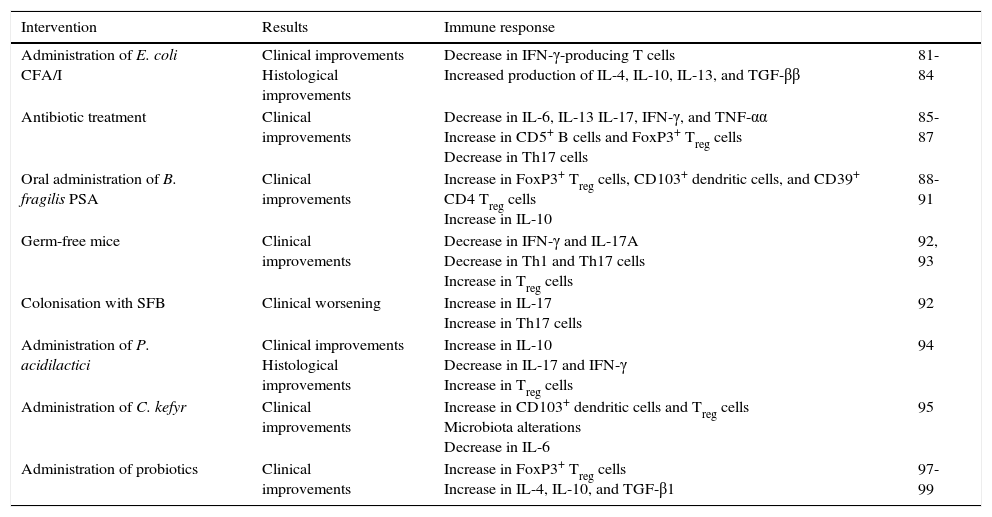
Summary of studies of the microbiota in experimental autoimmune encephalomyelitis.
| Intervention | Results | Immune response | |
|---|---|---|---|
| Administration of E. coli CFA/I | Clinical improvements Histological improvements | Decrease in IFN-γ-producing T cells Increased production of IL-4, IL-10, IL-13, and TGF-ββ | 81-84 |
| Antibiotic treatment | Clinical improvements | Decrease in IL-6, IL-13 IL-17, IFN-γ, and TNF-αα Increase in CD5+ B cells and FoxP3+ Treg cells Decrease in Th17 cells | 85-87 |
| Oral administration of B. fragilis PSA | Clinical improvements | Increase in FoxP3+ Treg cells, CD103+ dendritic cells, and CD39+ CD4 Treg cells Increase in IL-10 | 88-91 |
| Germ-free mice | Clinical improvements | Decrease in IFN-γ and IL-17A Decrease in Th1 and Th17 cells Increase in Treg cells | 92, 93 |
| Colonisation with SFB | Clinical worsening | Increase in IL-17 Increase in Th17 cells | 92 |
| Administration of P. acidilactici | Clinical improvements Histological improvements | Increase in IL-10 Decrease in IL-17 and IFN-γ Increase in Treg cells | 94 |
| Administration of C. kefyr | Clinical improvements | Increase in CD103+ dendritic cells and Treg cells Microbiota alterations Decrease in IL-6 | 95 |
| Administration of probiotics | Clinical improvements | Increase in FoxP3+ Treg cells Increase in IL-4, IL-10, and TGF-β1 | 97-99 |
Unlike studies of experimental mouse models, research on the gut microbiota in MS, that is, in humans, is still in early stages. The literature consists mainly of presentations at conferences,102 indirect evidence, and a few studies with small sample sizes (Table 3).
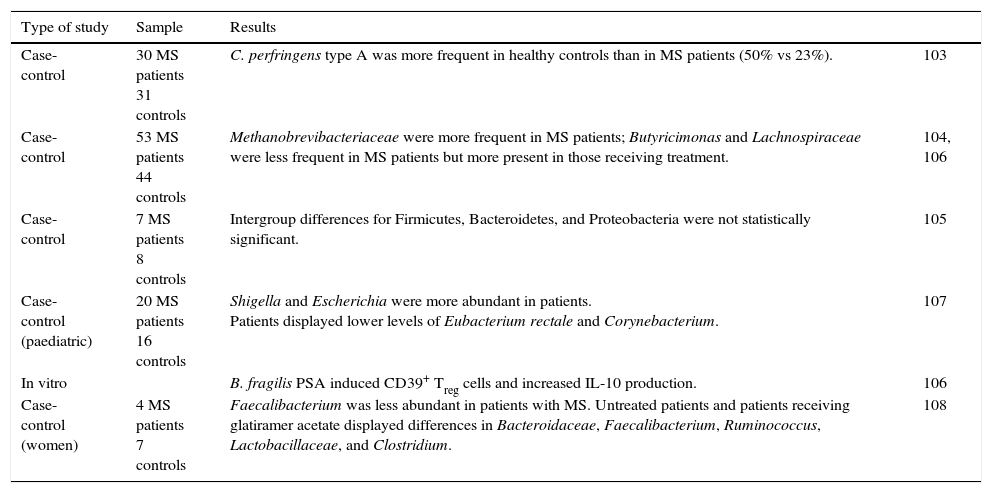
Summary of studies on the gut microbiota in MS.
| Type of study | Sample | Results | |
|---|---|---|---|
| Case-control | 30 MS patients 31 controls | C. perfringens type A was more frequent in healthy controls than in MS patients (50% vs 23%). | 103 |
| Case-control | 53 MS patients 44 controls | Methanobrevibacteriaceae were more frequent in MS patients; Butyricimonas and Lachnospiraceae were less frequent in MS patients but more present in those receiving treatment. | 104, 106 |
| Case-control | 7 MS patients 8 controls | Intergroup differences for Firmicutes, Bacteroidetes, and Proteobacteria were not statistically significant. | 105 |
| Case-control (paediatric) | 20 MS patients 16 controls | Shigella and Escherichia were more abundant in patients. Patients displayed lower levels of Eubacterium rectale and Corynebacterium. | 107 |
| In vitro | B. fragilis PSA induced CD39+ Treg cells and increased IL-10 production. | 106 | |
| Case-control (women) | 4 MS patients 7 controls | Faecalibacterium was less abundant in patients with MS. Untreated patients and patients receiving glatiramer acetate displayed differences in Bacteroidaceae, Faecalibacterium, Ruminococcus, Lactobacillaceae, and Clostridium. | 108 |
One trial analysed the association between MS and epsilon toxin-secreting bacillus C. perfringens type B, a non-commensal microorganism in humans; according to this study, commensal C. perfringens type A was more frequent in healthy controls than in patients with MS (50% vs 23%).103
Two studies addressing the role of the microbiota in MS were presented in poster format at the 2014 Annual Meeting of the American Academy of Neurology (AAN). Jhangi et al.104 found greater presence of Methanobrevibacteriaceae (Archaea) in patients with MS than in healthy controls. This microorganism has been associated with inflammatory processes and lower numbers of 2 potentially anti-inflammatory butyrate-producing microorganisms: the Butyricimonas genus from the Bacteroidetes phylum and the Lachnospiraceae family from the Firmicute phylum. In turn, their numbers were higher in MS patients receiving treatment. The second study found differences in populations of Firmicutes, Bacteroidetes, and Proteobacteria between patients with MS and controls, although these differences were not statistically significant (the sample included 7 patients and 8 controls).105
During the joint ACTRIMS-ECTRIMS Meeting held in Boston in 2014, 8 posters were presented in the “microbiome” category, including basic research studies, experimental studies in animals, and 4 studies in humans.106 The first of the studies conducted in humans included 20 paediatric patients and 16 age- and sex-matched controls. The MS patient group displayed significantly higher levels of Shigella and Escherichia, 2 bacteria associated with gastrointestinal infections, and lower levels of Eubacterium rectale and Corynebacterium.107 The second of these posters, presented by Jhangi et al., aimed to determine whether there were differences in the gut microbiota of patients with MS and whether MS was associated with a particular immune cell phenotype. In addition to the findings these researchers had reported in the poster presented at the 2014 AAN Annual Meeting,104 they suggested an association between Archaea and both IFN-γ expression and an activated phenotype of antigen-presenting cells in MS patients.
The authors of the third poster, including Ochoa-Repáraz, cultured antigen-presenting cells and T cells in the presence of PSA and found that PSA induced CD39+HLA-DR+FoxP3+ Treg cells and increased IL-10 production, and that PSA-induced Treg cells were more capable of suppressing TNF-α than control cells were. The last of the posters reporting on human studies was presented by the MS Microbiome Consortium, a multidisciplinary group aimed at determining the role of the gut microbiota in MS.
A small pilot study published in 2015 aimed to demonstrate the usefulness of vitamin D3 supplements in the treatment of MS.108 The study recruited 15 women (8 healthy controls and 7 patients with MS); 2 patients were untreated and 5 were receiving glatiramer acetate (3 of the latter did not submit stool samples in proper time/manner). Stool samples were collected at baseline and after vitamin D3 supplementation. Despite its small sample size, the study revealed differences in bacterial communities between patients with MS and controls. Faecalibacterium, a genus of bacteria with anti-inflammatory properties and associated with intestinal inflammatory disease, was less abundant in patients with MS and increased after vitamin D3 supplementation in patients who were not receiving glatiramer acetate. This study also found differences in community composition (including Bacteroidaceae, Faecalibacterium, Ruminococcus, Lactobacillaceae, and Clostridium) between untreated patients and patients receiving glatiramer acetate; these differences may be linked to immune response modulation by the drug.
ConclusionsThere is increasing evidence that the gut microbiota plays a major role in multiple functions of the human body, especially immune response. As a result, numerous studies have investigated the role of the intestinal flora in health and disease, especially autoimmune diseases.
In the field of MS, solid scientific evidence points to gut microbiota involvement in the pathogenesis, prevention, and treatment of EAE, the most widely-accepted animal model for MS. Some case-control studies extrapolating these results to MS have recently been published.
This promising new line of MS research should aim to identify the bacterial communities associated with MS, determine their role in the pathogenesis of the disease, and analyse the therapeutic potential of these findings.
Conflicts of interestThe authors have no conflicts of interest to declare.
Please cite this article as: Castillo-Álvarez F, Marzo-Sola M. Papel de la microbiota intestinal en el desarrollo de la esclerosis múltiple. Neurología. 2017;32:175–184.