The applications of artificial intelligence, and in particular automatic learning or “machine learning” (ML), constitute both a challenge and a great opportunity in numerous scientific, technical, and clinical disciplines. Specific applications in the study of multiple sclerosis (MS) have been no exception, and constitute an area of increasing interest in recent years.
ObjectiveWe present a systematic review of the application of ML algorithms in MS.
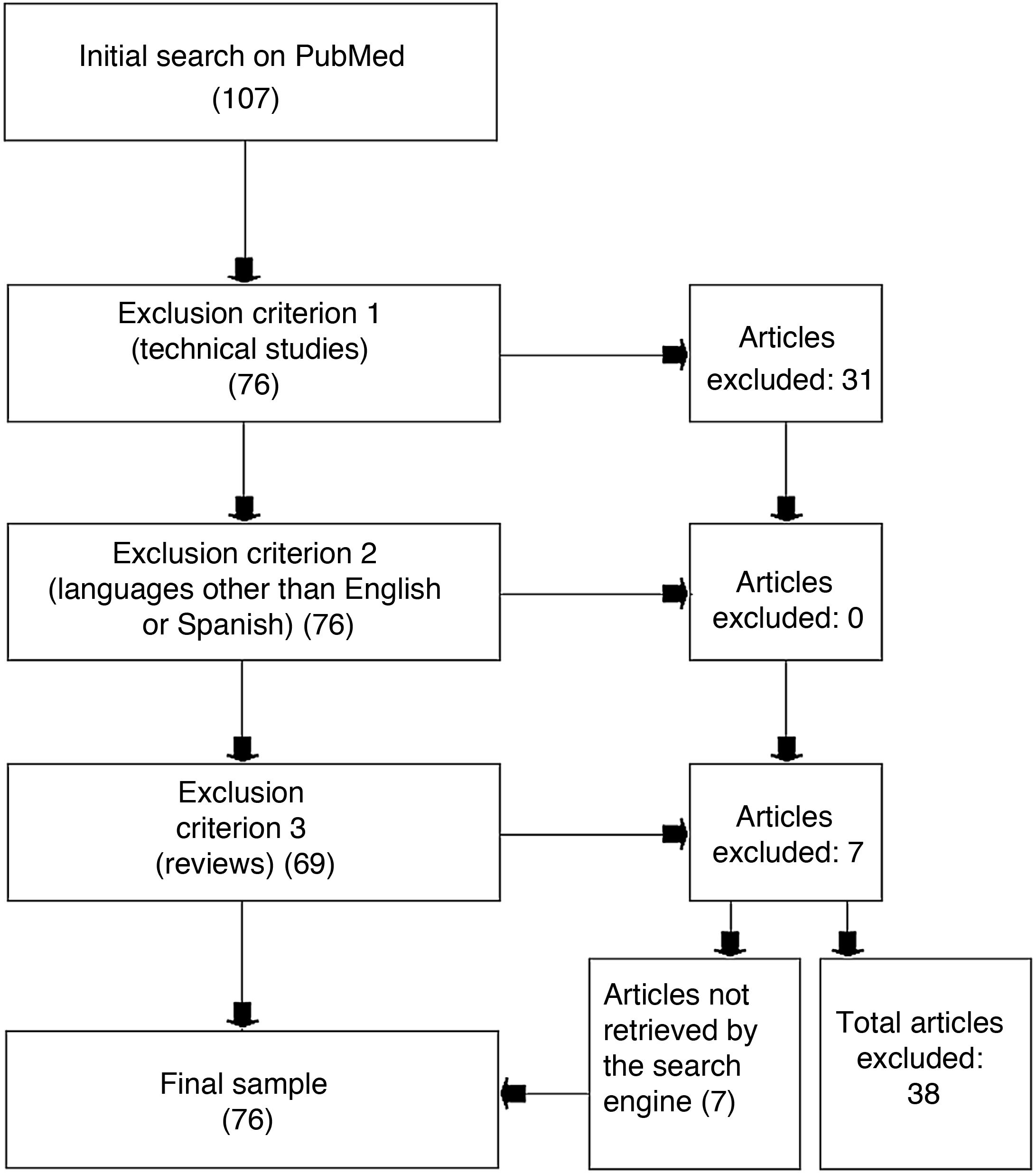
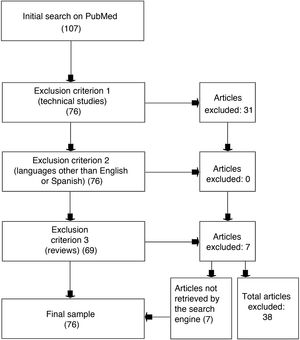
Materials and methodsWe used the PubMed search engine, which allows free access to the MEDLINE medical database, to identify studies including the keywords “machine learning” and “multiple sclerosis.” We excluded review articles, studies written in languages other than English or Spanish, and studies that were mainly technical and did not specifically apply to MS. The final selection included 76 articles, and 38 were rejected.
ConclusionsAfter the review process, we established 4 main applications of ML in MS: 1) classifying MS subtypes; 2) distinguishing patients with MS from healthy controls and individuals with other diseases; 3) predicting progression and response to therapeutic interventions; and 4) other applications. Results found to date have shown that ML algorithms may offer great support for health professionals both in clinical settings and in research into MS.
La aplicación de la inteligencia artificial y en particular de algoritmos de aprendizaje automático o “machine learning” (ML) constituye un desafío y al mismo tiempo una gran oportunidad en diversas disciplinas científicas, técnicas y clínicas. Las aplicaciones específicas en el estudio de la esclerosis múltiple (EM) no han sido una excepción mostrando un creciente interés en los últimos años.
ObjetivoRealizar una revisión sistemática de la aplicación de algoritmos de ML en la EM.
Material y métodosEmpleando el motor de búsqueda de libre acceso PubMed que accede a la base de datos MEDLINE, se seleccionaron aquellos estudios que incluyeran simultáneamente los dos siguientes conceptos de búsqueda: «machine learning» y «multiple sclerosis». Se rechazaron aquellos estudios que fueran revisiones, estuvieran en otro idioma que no fuera el castellano o el inglés, y aquellos trabajos que tuvieran un carácter técnico y no fueran aplicados para la esclerosis múltiple. Se seleccionaron como válidos setenta y seis artículos y fueron rechazados treinta y ocho.
ConclusionesTras la revisión de los estudios seleccionados, se pudo observar que la aplicación del ML en la EM se concentró en cuatro categorías: 1) clasificación de subtipos de pacientes dentro de la enfermedad; 2) diagnóstico del paciente frente a controles sanos u otras enfermedades; 3) predicción de la evolución o de la respuesta a intervenciones terapéuticas y por último 4) otros enfoques. Los resultados hallados hasta la fecha muestran que los diferentes algoritmos de ML pueden ser un gran apoyo para el profesional sanitario tanto en la clínica como en la investigación de la EM.
Artificial intelligence, and particularly machine learning (ML), are becoming increasingly widespread in numerous disciplines, with medicine being no exception. The past decade has seen a significant increase in the use and application of ML in the study of neurological diseases. This review analyses the first applications of ML in multiple sclerosis (MS), a disease with a considerable impact on society and healthcare.1 Before delving into the topic, we should point out that the present review does not seek to analyse the mathematical techniques underlying the ML algorithms used in different studies (see Nagy2 for a detailed review of this subject). However, we will discuss several concepts linked to ML that are frequently used in the studies included in this review. Artificial intelligence and ML are frequently confused. Machine learning is a field of artificial intelligence that focuses on the study of algorithms that learn from experience (data) to improve their performance in a specific task. For example, classification and regression tasks reveal patterns, which may be extremely complex, from the dataset of the study sample. ML may be approached from 2 different perspectives: supervised learning and unsupervised learning. In supervised learning, the algorithm is trained with a dataset labelled by the researcher, for example by disease subtype (eg, relapsing-remitting MS, secondary progressive MS, etc). This procedure takes on a predictive approach, with the algorithm defining the possible patterns of each label based on some or all the variables included in the analysis. The most relevant feature of ML in this field is that the algorithm is subsequently used to identify which of the previously defined patterns (learnt by the algorithm) best fit new data (eg, from a new patient). Unsupervised learning processes, in contrast, focus on descriptive tasks: the algorithm learns from unlabelled data, seeking similarities between data and establishing possible clusters or groups within the dataset. Readers may be aware of many supervised learning procedures, including neural networks, random forest, and support vector machine, as well as such unsupervised learning procedures as hierarchical clustering and self-organising maps.3 One of the main advantages of ML algorithms is that they can be used to analyse both quantitative (scores) and qualitative data (assessment by a healthcare professional) during the algorithmic learning process. Another key consideration is whether the ML algorithm is able to effectively classify patterns in the data. Several indicators are used to determine this, including accuracy, sensitivity, and specificity. Unfortunately, not all the studies included in this review include all these measures, and some even develop specific indicators, which hinders comparisons. This review presents an overview of the applications of ML in MS from the perspective of the interests and challenges of healthcare professionals. We also provide data on the algorithms’ accuracy in classifying patients.
Material and methodsWe used the free-access PubMed search engine to gather studies from MEDLINE, using the search terms “machine learning” and “multiple sclerosis,” combined with the Boolean operator AND.
We selected studies published until 30 April 2020 and applied the following exclusion criteria: 1) technical studies and studies not describing specific applications of ML to MS, 2) studies in languages other than Spanish or English, and 3) reviews. However, the reference lists of review studies were used to identify relevant studies that were not detected in the initial search.
The selection criteria involve a low risk of bias, although we also took measures to minimise this risk. For instance, we did not establish limitations on the type of journal (eg, quartile, impact factor) or the year of publication. Furthermore, we excluded review articles (to avoid duplicate articles), but did use them to identify studies not retrieved by the search engine. Lastly, we did not include studies focused on improving MRI studies, although we do make some remarks on this topic in the results section.
ResultsThe literature search identified 107 studies: 31 of these were excluded due to meeting the first exclusion criterion, none of the studies was excluded as per the second exclusion criterion, and 7 studies were excluded due to meeting the third exclusion criterion. An analysis of the reference lists of the 7 excluded review studies yielded a further 7 studies that had not been identified in the initial literature search; these were also included in our review. The review finally included a total of 76 studies and excluded 38 (Fig. 1).
The reviews identified in our literature search reveal a certain degree of maturity of the field of ML in neurology. However, most of them address multiple neurological diseases rather than focusing exclusively on MS.3–7 Only 2 reviews focus primarily on MS. One of these analyses the development of potential MS biomarkers based on MR spectroscopy data,8 while the other describes several applications of digital tools in MS and includes a section on the application of ML algorithms in these patients.9 Many studies analyse the suitability of ML algorithms for performing automated analysis of MRI studies. However, given the breadth of the topic addressed in this literature review, we will not analyse these studies in detail; readers interested in this topic can consult the articles included in the reference list.10–21 In any case, these studies aim to quantify whether ML algorithms can perform automatic classification of lesions with sufficient accuracy. The available data suggest that the application of these algorithms does not achieve sufficient accuracy, with the participation of healthcare professionals being essential. According to some studies, the critical aspect of the application of ML is not the algorithm used but rather the MRI parameters.11 Future studies are needed to establish valid procedures for the clinical application of ML in the identification of lesions on MR images.
The studies selected may be classified into 4 categories: classification of disease subtypes, diagnosis, prediction, and other approaches.
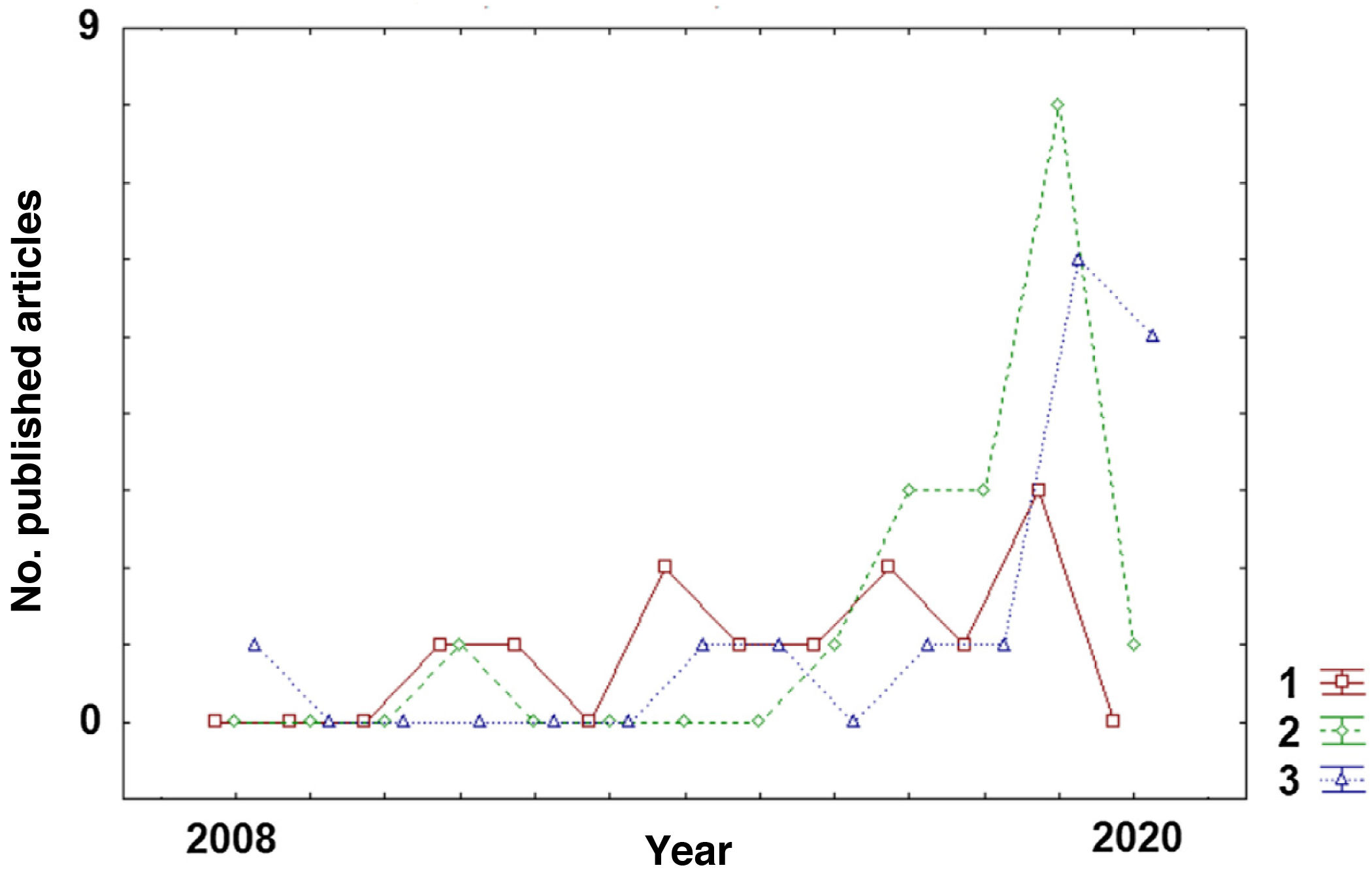
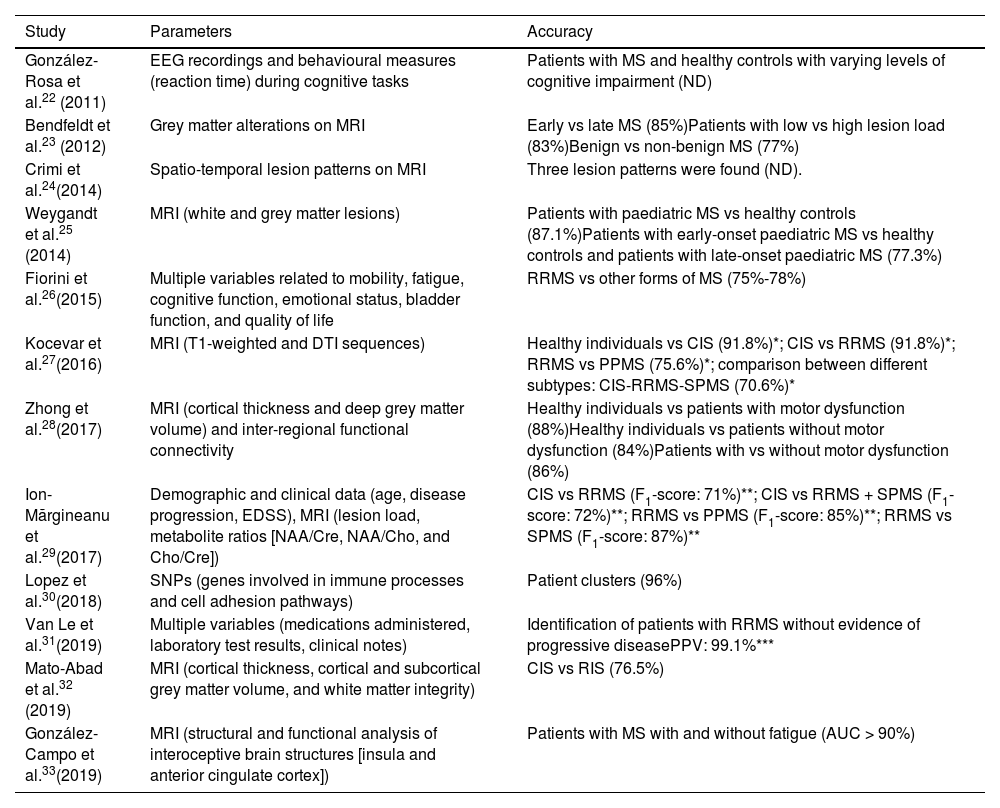
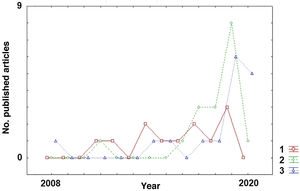
Classification of disease subtypesAlthough ML algorithms have only very recently begun to be applied to MS, there has been a clear shift in the aims of this technology (Fig. 2). Early studies aimed to classify patients by disease subtype. As shown in Table 1, studies with this objective22–33 were published more homogeneously over the past decade than studies with other objectives (diagnosis or prediction of MS) (Tables 2 and 3), which have increased considerably in recent years (2017-2020).
Studies into the application of machine learning algorithms for the classification of multiple sclerosis subtypes.
| Study | Parameters | Accuracy |
|---|---|---|
| González-Rosa et al.22 (2011) | EEG recordings and behavioural measures (reaction time) during cognitive tasks | Patients with MS and healthy controls with varying levels of cognitive impairment (ND) |
| Bendfeldt et al.23 (2012) | Grey matter alterations on MRI | Early vs late MS (85%)Patients with low vs high lesion load (83%)Benign vs non-benign MS (77%) |
| Crimi et al.24(2014) | Spatio-temporal lesion patterns on MRI | Three lesion patterns were found (ND). |
| Weygandt et al.25 (2014) | MRI (white and grey matter lesions) | Patients with paediatric MS vs healthy controls (87.1%)Patients with early-onset paediatric MS vs healthy controls and patients with late-onset paediatric MS (77.3%) |
| Fiorini et al.26(2015) | Multiple variables related to mobility, fatigue, cognitive function, emotional status, bladder function, and quality of life | RRMS vs other forms of MS (75%-78%) |
| Kocevar et al.27(2016) | MRI (T1-weighted and DTI sequences) | Healthy individuals vs CIS (91.8%)*; CIS vs RRMS (91.8%)*; RRMS vs PPMS (75.6%)*; comparison between different subtypes: CIS-RRMS-SPMS (70.6%)* |
| Zhong et al.28(2017) | MRI (cortical thickness and deep grey matter volume) and inter-regional functional connectivity | Healthy individuals vs patients with motor dysfunction (88%)Healthy individuals vs patients without motor dysfunction (84%)Patients with vs without motor dysfunction (86%) |
| Ion-Mărgineanu et al.29(2017) | Demographic and clinical data (age, disease progression, EDSS), MRI (lesion load, metabolite ratios [NAA/Cre, NAA/Cho, and Cho/Cre]) | CIS vs RRMS (F1-score: 71%)**; CIS vs RRMS + SPMS (F1-score: 72%)**; RRMS vs PPMS (F1-score: 85%)**; RRMS vs SPMS (F1-score: 87%)** |
| Lopez et al.30(2018) | SNPs (genes involved in immune processes and cell adhesion pathways) | Patient clusters (96%) |
| Van Le et al.31(2019) | Multiple variables (medications administered, laboratory test results, clinical notes) | Identification of patients with RRMS without evidence of progressive diseasePPV: 99.1%*** |
| Mato-Abad et al.32 (2019) | MRI (cortical thickness, cortical and subcortical grey matter volume, and white matter integrity) | CIS vs RIS (76.5%) |
| González-Campo et al.33(2019) | MRI (structural and functional analysis of interoceptive brain structures [insula and anterior cingulate cortex]) | Patients with MS with and without fatigue (AUC > 90%) |
AUC: area under the curve; CIS: clinically isolated syndrome; EDSS: Expanded Disability Status Scale; EEG: electroencephalography; MRI: magnetic resonance imaging; MS: multiple sclerosis; ND: not disclosed; PPMS: primary progressive multiple sclerosis; PPV: positive predictive value; RIS: radiologically isolated syndrome; RRMS: relapsing-remitting multiple sclerosis; SNP: single nucleotide polymorphism; SPMS: secondary progressive multiple sclerosis.
Values correspond to what the authors term the “F-measure,” which combines precision and recall (proportion of true positives identified among false negatives). See the study by Kocevar et al.27 for a definition of the formula.
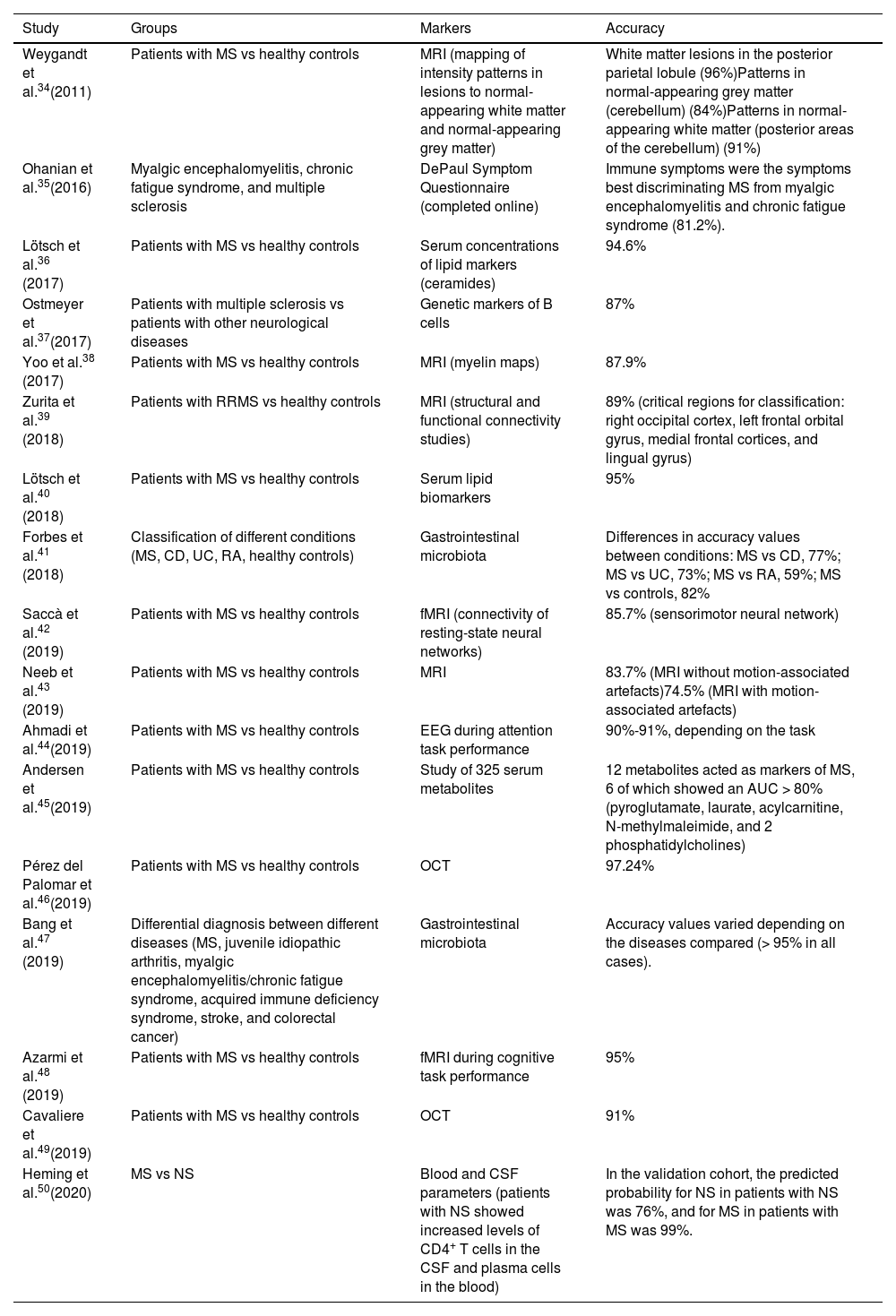
Studies into the application of machine learning algorithms for diagnosing multiple sclerosis.
| Study | Groups | Markers | Accuracy |
|---|---|---|---|
| Weygandt et al.34(2011) | Patients with MS vs healthy controls | MRI (mapping of intensity patterns in lesions to normal-appearing white matter and normal-appearing grey matter) | White matter lesions in the posterior parietal lobule (96%)Patterns in normal-appearing grey matter (cerebellum) (84%)Patterns in normal-appearing white matter (posterior areas of the cerebellum) (91%) |
| Ohanian et al.35(2016) | Myalgic encephalomyelitis, chronic fatigue syndrome, and multiple sclerosis | DePaul Symptom Questionnaire (completed online) | Immune symptoms were the symptoms best discriminating MS from myalgic encephalomyelitis and chronic fatigue syndrome (81.2%). |
| Lötsch et al.36 (2017) | Patients with MS vs healthy controls | Serum concentrations of lipid markers (ceramides) | 94.6% |
| Ostmeyer et al.37(2017) | Patients with multiple sclerosis vs patients with other neurological diseases | Genetic markers of B cells | 87% |
| Yoo et al.38 (2017) | Patients with MS vs healthy controls | MRI (myelin maps) | 87.9% |
| Zurita et al.39 (2018) | Patients with RRMS vs healthy controls | MRI (structural and functional connectivity studies) | 89% (critical regions for classification: right occipital cortex, left frontal orbital gyrus, medial frontal cortices, and lingual gyrus) |
| Lötsch et al.40 (2018) | Patients with MS vs healthy controls | Serum lipid biomarkers | 95% |
| Forbes et al.41 (2018) | Classification of different conditions (MS, CD, UC, RA, healthy controls) | Gastrointestinal microbiota | Differences in accuracy values between conditions: MS vs CD, 77%; MS vs UC, 73%; MS vs RA, 59%; MS vs controls, 82% |
| Saccà et al.42 (2019) | Patients with MS vs healthy controls | fMRI (connectivity of resting-state neural networks) | 85.7% (sensorimotor neural network) |
| Neeb et al.43 (2019) | Patients with MS vs healthy controls | MRI | 83.7% (MRI without motion-associated artefacts)74.5% (MRI with motion-associated artefacts) |
| Ahmadi et al.44(2019) | Patients with MS vs healthy controls | EEG during attention task performance | 90%-91%, depending on the task |
| Andersen et al.45(2019) | Patients with MS vs healthy controls | Study of 325 serum metabolites | 12 metabolites acted as markers of MS, 6 of which showed an AUC > 80% (pyroglutamate, laurate, acylcarnitine, N-methylmaleimide, and 2 phosphatidylcholines) |
| Pérez del Palomar et al.46(2019) | Patients with MS vs healthy controls | OCT | 97.24% |
| Bang et al.47 (2019) | Differential diagnosis between different diseases (MS, juvenile idiopathic arthritis, myalgic encephalomyelitis/chronic fatigue syndrome, acquired immune deficiency syndrome, stroke, and colorectal cancer) | Gastrointestinal microbiota | Accuracy values varied depending on the diseases compared (> 95% in all cases). |
| Azarmi et al.48 (2019) | Patients with MS vs healthy controls | fMRI during cognitive task performance | 95% |
| Cavaliere et al.49(2019) | Patients with MS vs healthy controls | OCT | 91% |
| Heming et al.50(2020) | MS vs NS | Blood and CSF parameters (patients with NS showed increased levels of CD4+ T cells in the CSF and plasma cells in the blood) | In the validation cohort, the predicted probability for NS in patients with NS was 76%, and for MS in patients with MS was 99%. |
AUC: area under the curve; CD: Crohn disease; CSF: cerebrospinal fluid; fMRI: functional magnetic resonance imaging; MRI: magnetic resonance imaging; MS: multiple sclerosis; NS: neurosarcoidosis; OCT: optic coherence tomography; RA: rheumatoid arthritis; RRMS: relapsing-remitting multiple sclerosis; UC: ulcerative colitis.
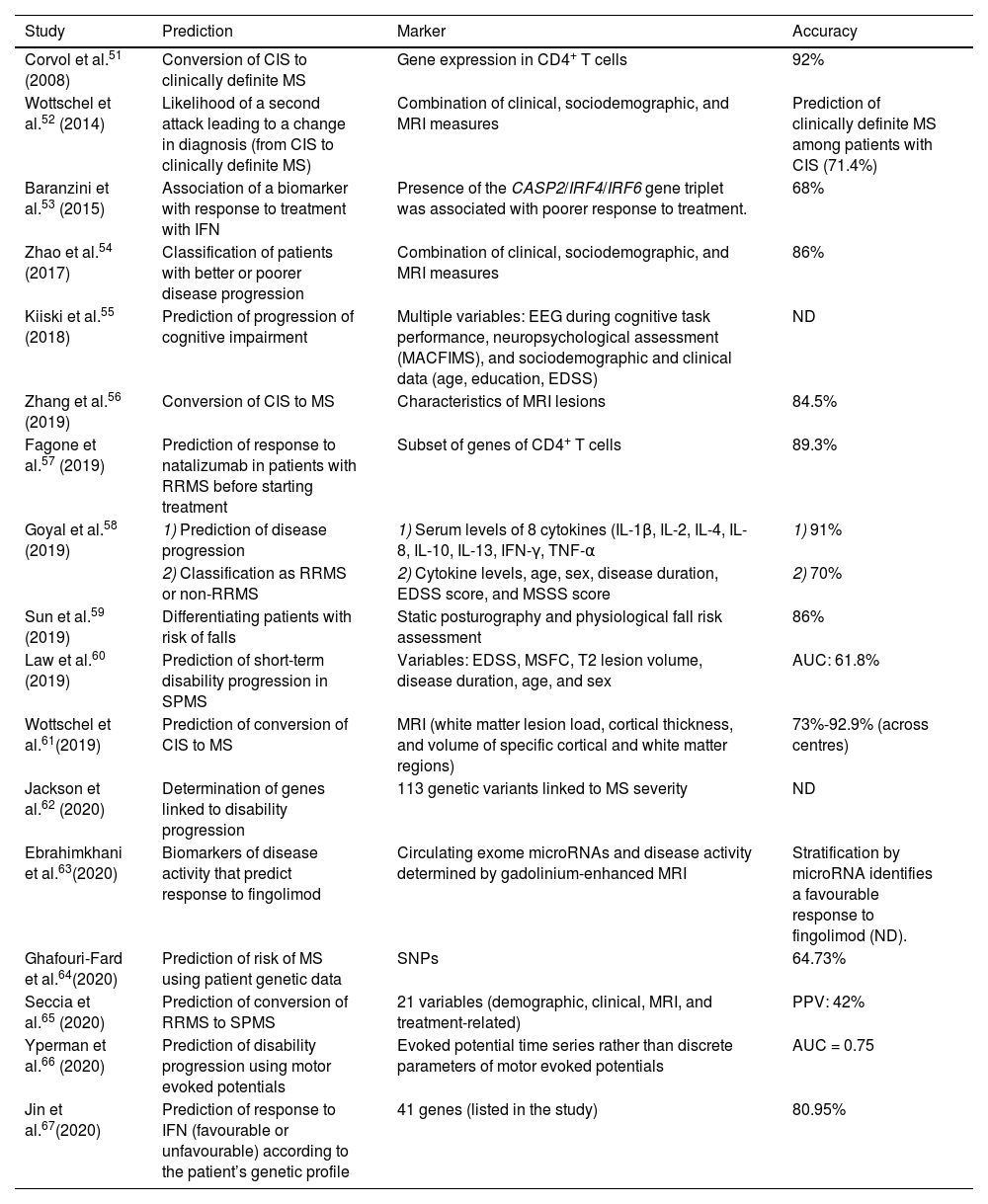
Studies into the application of machine learning algorithms for predicting disease progression or treatment response.
| Study | Prediction | Marker | Accuracy |
|---|---|---|---|
| Corvol et al.51 (2008) | Conversion of CIS to clinically definite MS | Gene expression in CD4+ T cells | 92% |
| Wottschel et al.52 (2014) | Likelihood of a second attack leading to a change in diagnosis (from CIS to clinically definite MS) | Combination of clinical, sociodemographic, and MRI measures | Prediction of clinically definite MS among patients with CIS (71.4%) |
| Baranzini et al.53 (2015) | Association of a biomarker with response to treatment with IFN | Presence of the CASP2/IRF4/IRF6 gene triplet was associated with poorer response to treatment. | 68% |
| Zhao et al.54 (2017) | Classification of patients with better or poorer disease progression | Combination of clinical, sociodemographic, and MRI measures | 86% |
| Kiiski et al.55 (2018) | Prediction of progression of cognitive impairment | Multiple variables: EEG during cognitive task performance, neuropsychological assessment (MACFIMS), and sociodemographic and clinical data (age, education, EDSS) | ND |
| Zhang et al.56 (2019) | Conversion of CIS to MS | Characteristics of MRI lesions | 84.5% |
| Fagone et al.57 (2019) | Prediction of response to natalizumab in patients with RRMS before starting treatment | Subset of genes of CD4+ T cells | 89.3% |
| Goyal et al.58 (2019) | 1) Prediction of disease progression | 1) Serum levels of 8 cytokines (IL-1β, IL-2, IL-4, IL-8, IL-10, IL-13, IFN-γ, TNF-α | 1) 91% |
| 2) Classification as RRMS or non-RRMS | 2) Cytokine levels, age, sex, disease duration, EDSS score, and MSSS score | 2) 70% | |
| Sun et al.59 (2019) | Differentiating patients with risk of falls | Static posturography and physiological fall risk assessment | 86% |
| Law et al.60 (2019) | Prediction of short-term disability progression in SPMS | Variables: EDSS, MSFC, T2 lesion volume, disease duration, age, and sex | AUC: 61.8% |
| Wottschel et al.61(2019) | Prediction of conversion of CIS to MS | MRI (white matter lesion load, cortical thickness, and volume of specific cortical and white matter regions) | 73%-92.9% (across centres) |
| Jackson et al.62 (2020) | Determination of genes linked to disability progression | 113 genetic variants linked to MS severity | ND |
| Ebrahimkhani et al.63(2020) | Biomarkers of disease activity that predict response to fingolimod | Circulating exome microRNAs and disease activity determined by gadolinium-enhanced MRI | Stratification by microRNA identifies a favourable response to fingolimod (ND). |
| Ghafouri-Fard et al.64(2020) | Prediction of risk of MS using patient genetic data | SNPs | 64.73% |
| Seccia et al.65 (2020) | Prediction of conversion of RRMS to SPMS | 21 variables (demographic, clinical, MRI, and treatment-related) | PPV: 42% |
| Yperman et al.66 (2020) | Prediction of disability progression using motor evoked potentials | Evoked potential time series rather than discrete parameters of motor evoked potentials | AUC = 0.75 |
| Jin et al.67(2020) | Prediction of response to IFN (favourable or unfavourable) according to the patient’s genetic profile | 41 genes (listed in the study) | 80.95% |
AUC: area under the curve; CIS: clinically isolated syndrome; EDSS: Expanded Disability Status Scale; IFN: interferon; MS: multiple sclerosis; MSFC: Multiple Sclerosis Functional Composite; MSSS: Multiple Sclerosis Severity Scale; ND: not disclosed; PPV: positive predictive value; RRMS: relapsing-remitting multiple sclerosis; SNP: single nucleotide polymorphism; SPMS: secondary progressive multiple sclerosis.
The available studies have enabled the classification of patients into several categories (relapsing-remitting MS, benign MS, clinically isolated syndrome, radiologically isolated syndrome, early- or late-onset paediatric MS), by specific characteristics (eg, lesion load, disease onset), and even by degree of cognitive impairment. Many variables are used for classification, from MRI parameters related to the grey and white matter to clinical measures (eg, EDSS, disease duration) and even qualitative variables, such as clinical notes.
The accuracy achieved by these studies ranges from 70.6% to 96%. Accuracy values vary greatly between studies, which suggests that accuracy is strongly influenced by the algorithms applied, the variables used for classification, and the disease subtypes the study aims to identify. We may therefore conclude that ML algorithms can achieve acceptable levels of accuracy in classifying patients by disease subtype, and may improve the stratification of these patients for subsequent studies into response to treatment interventions or disease progression.
DiagnosisAccording to our literature search, the first study into the application of ML algorithms for the diagnosis of MS was published in 2011; no further studies were published until 2016. Interestingly, a large proportion of the studies included in this category were published in 2019 and, unlike in the case of studies into the prediction of MS, only one study was published in the first 4 months of 2020 (Table 2).34–50 Furthermore, studies in this category mainly focus on the diagnosis of MS as compared to healthy controls (13 studies), while fewer studies (5 studies) aimed to differentiate MS from other diseases.
The main variables used to improve diagnostic ability are derived from different MRI parameters (lesion maps, functional connectivity, etc). However, other markers offer interesting levels of accuracy, including serological and genetic markers, or such techniques as optical coherence tomography (OCT) or EEG.
The level of accuracy of these markers ranges from very good (98.2%) to moderate (59%). Curiously, these 2 percentages correspond to studies comparing the characteristics of the gut microbiota between patients with MS and patients with other conditions. One of the studies aimed to differentiate MS from such other conditions as idiopathic arthritis and chronic fatigue syndrome,47 whereas the other achieved a considerably lower level of accuracy when comparing MS against rheumatoid arthritis.40
PredictionThis section includes several studies that essentially classify patients by disease subtype. This is the case with studies aiming to discriminate between favourable and unfavourable response to pharmacological treatment. The decision to include these studies in this category was based on the fact that the ultimate goal of applying an ML algorithm is not only classification but rather prediction.
The first study into predicting progression was published in 2008; several years passed before additional studies with the same approach were published, in 2014 (Table 3).51–67 However, this is currently the approach that has been most explored in the past 2 years, with 6 studies being published in the first 4 months of 2020. The different lines of research in this field have 2 main goals. On the one hand, they seek to predict disease progression (eg, risk of progression to a more aggressive disease course), and on the other, they aim to predict patient response to a pharmacological treatment to assist healthcare professionals in selecting the most appropriate treatment. This approach undoubtedly contributes to more personalised care, as it acknowledges patient heterogeneity. The accuracy of ML algorithms in predicting disease progression ranged from 61.8% to 92.9%, whereas the accuracy in classifying patients by response to pharmacological treatment ranged from 68% to 89.3%.
Other approachesThis last section focuses on other lines of research into the application of ML in MS. One of the most active lines of research is the development of new indicators (scores) integrating different variables and improving patient assessment. One example of this application is the algorithm developed by Kosa et al.68 in 2016. This research group underscored the need to obtain an indicator with greater sensitivity to changes in disease progression, with a view to predicting responses to pharmacological treatment in patients with progressive forms of the disease. This new outcome measure, known as CombiWISE, integrates 4 scales: the Expanded Disability Status Scale (EDSS), the Scripps neurological rating scale, the Timed 25-Foot Walk, and the 9-Hole Peg Test. CombiWISE was used to follow up 98 patients with progressive MS, and was found to be superior to MRI atrophy measurement and the EDSS. In fact, a 1-point change in EDSS scores corresponded to a 7.5-point change on CombiWISE, with the latter instrument therefore showing greater resolution for patient assessment. Another research group69 developed an ML algorithm to calculate an indicator termed brain age gap and evaluated its usefulness as a marker of clinical course and severity. According to their results, this indicator showed a higher mean brain age in patients with MS than in healthy controls, and was found to be correlated with brain atrophy and white matter lesion load. Unfortunately, the correlation between brain age gap and several clinical variables was poor, raising questions about its usefulness in the clinical setting. In other cases, ML algorithms have focused on optimising assessment with existing tools. A very active area of research is the evaluation of motor disability and its association with several parameters. These studies frequently assess gait using skin-mounted wearable sensors.70–73 Another active line of research is the analysis of potential causes of the disease. The purpose of these studies is twofold: describing possible risk factors74,75 and identifying potential markers of MS with a view to developing therapeutic targets for future treatments.76–79 Another interesting line of research uses different approaches to evaluate whether ML is beneficial for healthcare professionals from a purely practical standpoint. Some studies have evaluated whether the application of ML algorithms improves the prediction of disease progression. One study aimed to predict disease course based on patient clinical records.80 The study concluded that cooperation between healthcare professionals and ML algorithms yielded better predictions than those of healthcare professionals or ML algorithms alone.
The application of algorithms reduces the impact of confounding factors on disease classification. One study demonstrated that an ML algorithm can minimise the impact of such confounding factors as age, which can reduce the homogeneity of study groups (clusters), without the intervention of a healthcare professional.81 In other studies, the application of ML improves test application by reducing operation times, as is the case with OCT, where ML achieves more precise segmentation of the retinal layer and macular oedema than other more time-consuming methods, as the segmentation operation only takes 10 seconds.82 Another application of ML to support healthcare professionals, in this case neuropsychologists, is the partial automation of cognitive assessments.83 Cognitive impairment may appear over the course of MS. One of the tests used for cognitive assessment is the Brief Visuospatial Memory Test-Revised, in which patients have to identify and draw geometric figures. After digitalising the test and training the algorithm, the authors obtained 80% accuracy in interpreting test results, using professional assessment as the gold standard. They concluded that the initial assessment must still be performed by a healthcare professional, but improvements in learning algorithms may allow ML algorithms to reliably reassess cognitive function in longitudinal studies.
DiscussionWithout a doubt, the most intuitive applications of ML algorithms in MS are in diagnosis (or classification by disease subtype) and in the prediction of disease progression or treatment response. However, ML algorithms have a wide range of applications, and recent studies show multiple new lines of research. In the case of MS, these algorithms enable the analysis of vast amounts of omics data; until very recently, this was extremely difficult with conventional statistical analysis techniques. Particularly interesting is the application of ML algorithms to the study of MS biomarkers, which may help us to determine disease aetiology and to establish future treatment strategies.
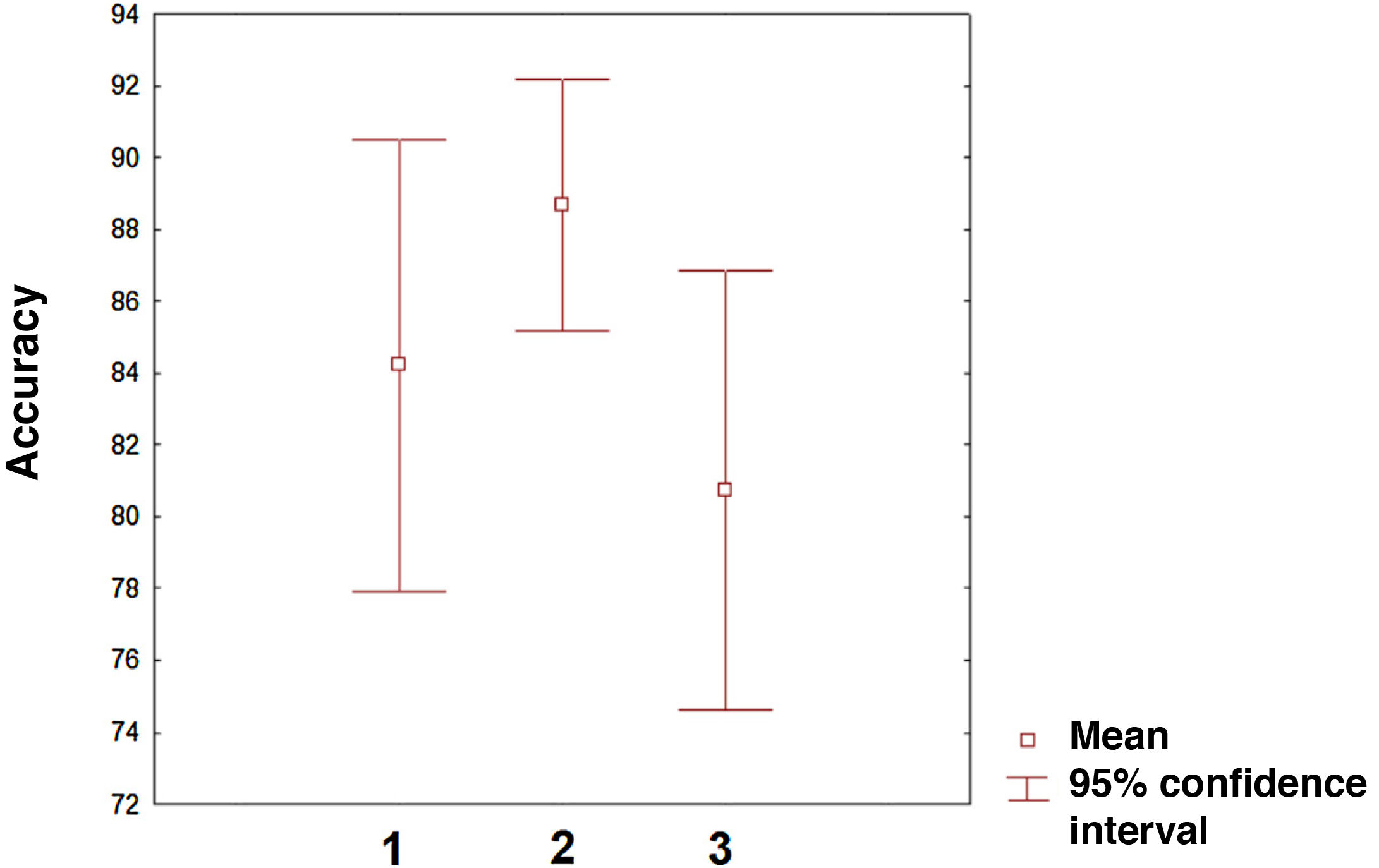
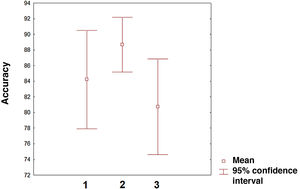
The field with the greatest potential for clinical application of ML algorithms is the prediction of treatment response to new and existing therapies. In fact, prediction is the application on which the greatest number of studies have been published in the last 2 years. However, there is still an imbalance between the studies applying algorithms to predict disease progression and a minority of studies focusing on predicting treatment response. In the near future, the latter line of research will surely become the most common application of ML in MS and other neurological diseases. The 3 categories analysed (classification, diagnosis, and prediction) present similar accuracy, although confidence intervals vary (Fig. 3). Comparisons between categories should be made with caution given that measures of accuracy, the number of variables included, and the ML procedures used vary between studies. In any case, we may conclude that the first studies in this field show considerable accuracy. Given the training needed by these algorithms, there is a need for healthcare institutions to cooperate through shared data repositories, with a view to increasing the ability of these algorithms to describe disease patterns. This approach is particularly interesting in the case of genetic data, as it will enable more accurate characterisation of patients and more personalised treatments. Unquestionably, we are on the path toward personalised medicine, as suggested by multiple studies.84–86 In the coming years, healthcare systems that make the effort to implement artificial intelligence tools will surely lead some of the most relevant advances in clinical practice. Our study is not without limitations. The PubMed search engine may have missed some relevant articles. However, analysis of the reference lists of the articles analysed minimises the possibility that we failed to include a large number of articles on ML and MS. Furthermore, other concepts related to ML, such as deep learning, may have interesting applications in MS. Therefore, future reviews on the application of ML in MS or other diseases should include such other keywords as deep learning or network representation learning.
ConclusionThis review shows that the application of ML algorithms in MS is an active field that has grown exponentially in the past 2 years. The main applications for this promising technology are in the classification of disease subtypes, diagnosis of MS as compared to healthy individuals or patients with other conditions, and prediction of disease progression or treatment response. However, some studies have also focused on optimising techniques or seeking new indicators with a view to improving clinical assessment in terms of time, cost, and patient well-being. In general terms, the published evidence suggests that the application of ML algorithms achieves excellent accuracy (> 90% in some studies), and may represent an important tool for healthcare professionals in the diagnosis and prognosis of MS.
FundingThis study was funded by the Spanish Ministry of Science, Innovation, and Universities (project code PSI2016-78133-P).
Conflicts of interestThe authors have no conflicts of interest to declare.