Pseudomonas syringae pv. phaseolicola is a phytopathogenic bacterium in beans that produces a phytotoxin called phaseolotoxin, in whose synthesis a group of genes that belong to the “Pht cluster” are involved. This cluster comprises 23 genes arranged in 5 transcriptional units, two monocistronic (argK, phtL) and three polycistronic (phtA, phtD, phtM) operons, whose expression is increased at 18°C, correlating with the production of phaseolotoxin by the bacterium. So far, the regulatory mechanisms involved in phaseolotoxin synthesis are poorly understood and only the requirement of low temperatures for its synthesis has been demonstrated. Therefore, in this study we searched for regulatory proteins that could be involved in the phaseolotoxin synthesis, focusing on the regulation of the phtM operon. Gel shift assays showed that the promoter region of the phtM operon contains binding sites for putative regulatory proteins, which are encoded outside the Pht cluster and are independent of the GacS–GacA two-component system. Deletion assays with the promoter region of the phtM operon show that the binding site for a putative transcription factor is located within a 58bp region. The putative transcription factor of the phtM operon has an apparent molecular mass in the 14–20kDa range. Furthermore, the results demonstrate that the transcription factor recognizes and binds the upstream phtM region as monomer o multimer of a single polypeptide. Our findings provide new insights into the regulatory mechanisms involved in phaseolotoxin production, and suggest that the Pht cluster was integrated into the global regulatory mechanism of P. syringae pv. phaseolicola.
Pseudomonas syringae pv. phaseolicola es una bacteria patógena del frijol que produce una fitotoxina conocida como faseolotoxina. En la síntesis de dicha molécula se encuentran involucrados un grupo de genes que conforman el denominado cluster Pht. Este cluster comprende 23 genes, ordenados en 5 unidades transcripcionales —2 monocistrónicas (argK, phtL) y 3 poliscistrónicas (phtA, phtD, phtM)—, cuya expresión se encuentra incrementada a 18°C, en correlación con la producción de faseolotoxina por la bacteria. Hasta ahora, los mecanismos regulatorios involucrados en la síntesis de faseolotoxina han sido poco estudiados. El objetivo de este trabajo fue buscar y caracterizar proteínas regulatorias involucradas en la síntesis de faseolotoxina, colocando el foco en la regulación del operón phtM. Los ensayos de cambio en la movilidad electroforética mostraron que la región promotora del operón phtM contiene sitios de unión para posibles proteínas regulatorias, codificadas fuera del cluster Pht e independientes del sistema de dos componentes GacS-GacA. El sitio de unión para estos factores de transcripción está localizado en una región de 58 pb. Los resultados demuestran, además, que el probable factor de transcripción del operón phtM tiene una masa molecular aparente de 14-20kDa y que reconoce y se une a la región río arriba de phtM como monómero o multímero de un solo polipéptido. Este estudio aporta nuevos hallazgos dentro de los mecanismos regulatorios involucrados en la síntesis de faseolotoxina, los que indican que el cluster Pht se integró a los mecanismos regulatorios de P. syringae pv. phaseolicola.
Phaseolotoxin [Nδ(N′-sulfodiaminophosphinyl)-ornithyl-alanyl-homoarginine], is an extracellular, nonhost-specific and chlorosis-inducing toxin, considered to be the major virulence factor of the phytopathogenic bacterium Pseudomonas syringae pv. phaseolicola, the causal agent of the disease in beans (Phaseolus vulgaris L.) known as “Halo Blight”32. This is considered one of the most common diseases of beans in temperate and cold regions, and is of economic importance in the world due to major field crop losses. The halo blight disease is characterized by the development of a chlorotic zone or halo around the necrotic infection site. This halo results from the action of phaseolotoxin released from the bacteria in the infection site and its diffusion into the surrounding leaf tissues21–23. Phaseolotoxin acts as reversible inhibitor of the ornithine carbamoyltransferase enzyme (OCTase; EC2.1.3.3) which catalyses the conversion of ornithine to citruline, a reaction common to both arginine biosynthesis and the urea cycle, thus resulting in host cell death. In addition, phaseolotoxin facilitates the systemic invasion of the plant, contributing significantly to the virulence of the pathogen11,34. In this manner, phaseolotoxin is a key element for halo blight disease development. However, non-phaseolotoxin-producing strains have also been detected in bean fields with epidemiological importance and causing the same lesions in the plants except the absence of chlorotic halos13,28. The production of phaseolotoxin by P. syringae pv. phaseolicola is mainly temperature-dependent, with high levels of this metabolite at 18°C–20°C, while no detectable amounts of the toxin are present at 28°C–30°C, corresponding to the optimal growth temperature for this bacterium15,25. Genes necessary for the synthesis of phaseolotoxin are encoded within a 30.24kb pathogenicity island (PAI) termed the “Pht cluster”, which contains 23 genes arranged into five transcriptional units, including two monocistronic (argK and phtL) and three polycistronic operons; one comprising 11 genes from phtA to phtK with an internal promoter driving expression of phtD to phtK and a third large polycistronic operon comprising 10 genes from phtM to phtV1,12,30,31. The function of only few of these genes has been elucidated; argK encoding a phaseolotoxin-insensitive OCTase, which provides resistance to the bacterium to its own toxin24,27; desI coding for a fatty acid desaturase16, amtA encoding an amidinotransferase17; phtU coding an L-amino acid ligase3 and phtL, whose product has a regulatory function on both the phaseolotoxin cluster (Pht) and genomic genes1,14. The presence of the Pht cluster has also been reported in the other known phaseolotoxin-producing pathovars P. syringae pv. actinidiae (kiwi pathogen) and in a single strain of P. syringae pv. syringae CFBP3388, although in the latter the cluster organization is poorly conserved33,35. So far, it is still unknown whether this cluster contains all the elements necessary for phaseolotoxin production. Recently, the PSPPH_4550 gene encoding for a putative non-ribosomal peptide synthetase, located outside the Pht cluster, has been identified and shown to be involved in the synthesis of phaseolotoxin in P. syringae pv. phaseolicola NPS3121. However, this gene appears to be related to bacterial fitness, in which the Pht cluster genes have been integrated after a presumed horizontal gene-transfer event9.
The regulatory mechanisms that control phaseolotoxin gene expression in P. syringae pv. phaseolicola are just beginning to be elucidated. The expression of Pht cluster genes occurs selectively during growth at low temperature (18°C), while at 28°C only basal levels of expression are observed for these genes. This is consistent with the permissive temperature for phaseolotoxin synthesis, with the exception of the expression of phtL which was detected at both temperatures1. The way P. syringae pv. phaseolicola regulates the expression of these genes and the synthesis of phaseolotoxin in relation to temperature is poorly understood. The molecular basis involved in the signal transduction pathway and/or regulatory networks that participate in this process have not been elucidated yet. The analyses of transcriptional fusions of intergenic regions of Pht cluster genes suggest that low temperature regulation occurs at the transcriptional level. Furthermore, analysis of the promoter regions identified in the Pht cluster showed that the divergent promoters for argK and phtA present a −35 and −10 region that is characteristic of the negatively controlled promoters, while the promoter regions for phtD, phtL and phtM did not show any similarity to consensus sequences for bacterial sigma factors. Although a common mechanism of transcriptional regulation for phtD and phtM has been suggested due to the presence of conserved regions in the promoters of these operons1. So far, the studies focused on the identification of regulatory proteins involved in the expression of Pht cluster genes are very scarce. A regulatory function for the PhtL protein has been suggested based on the lack of phtM operon expression in a phtL− background1. However this has not been confirmed yet. Similarly, a regulatory function for the phtABC genes has been proposed, participating in the transcriptional repression of the argK gene2. So far, only the IHF (Integration Host Factor) protein has been identified as involved, in a direct manner, in the regulation of Pht cluster genes, particularly in the phtD operon4. For these reasons, the goal of this study aims to delve into the regulatory pathways involved in the phaseolotoxin synthesis in P. syringae pv. phaseolicola by identifying and/or characterizing regulatory proteins that could participate in the regulation of the expression of Pht cluster genes by focusing on the phtM operon.
Materials and methodsBacterial strains, media and growth conditionsP. syringae strains:pv. phaseolicola NPS3121 wild type (wt), gacA− mutant, pv. phaseolicola CLY233, and pv. tomato DC3000 were grown on M9 minimal medium at 18°C or 28°C1,7,9,28. Pre-inocula (25ml) of P. syringae strains were grown overnight at 28°C in M9 medium with glucose (0.8%) as carbon source. The cells were inoculated into 50ml M9 minimal medium at OD600nm 0.1 and the cultures were incubated at 18°C or 28°C until they reached early stationary phase (OD600nm 1.0). When required, antibiotics were added to cultures of P. syringae pv. phaseolicola NPS3121wt (rifampin 50μg/ml) and gacA− mutant strains (rifampin 50μg/ml, kanamycin 70μg/ml).
Biochemistry and molecular biology techniquesGenomic DNA of P. syringae pv. phaseolicola NPS3121 was isolated as previously described8. Routine molecular techniques were performed using standard protocols29. PCR products were amplified with High Fidelity DNA Polymerase and Platinum supermix (Invitrogen, California USA) and purified with the QIAquick® gel extraction kit (QIAGEN, Hilden, Germany). Primers were designed using the Vector NTI Software (Invitrogen) on the basis of the previously reported Pht cluster sequence (GenBank DQ1412631) (Fig. 1A). The oligonucleotides used in this study are listed under supplementary material 1. The biochemical procedures such as SDS-PAGE and electroblotting were performed according to standard protocols29.
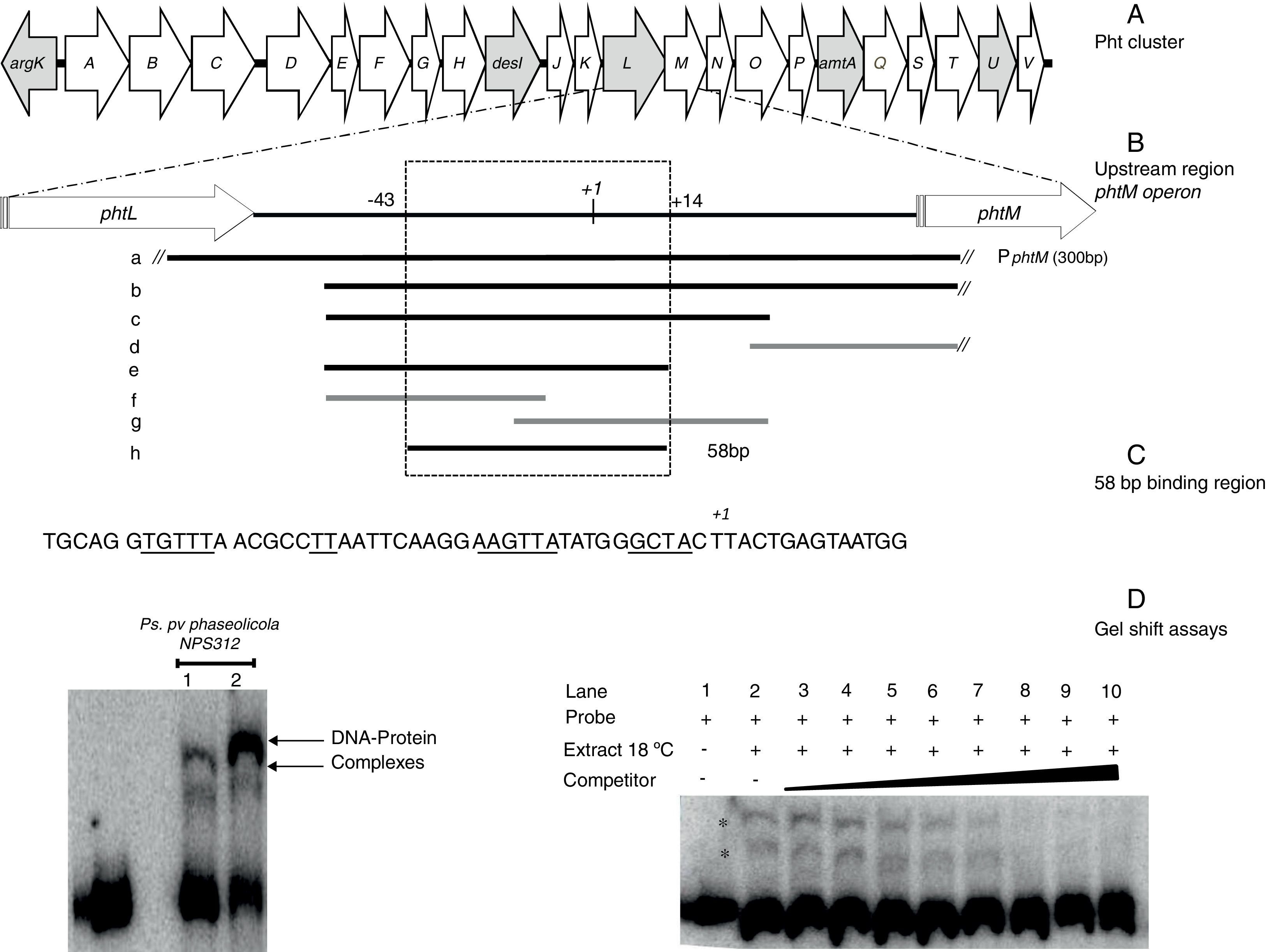
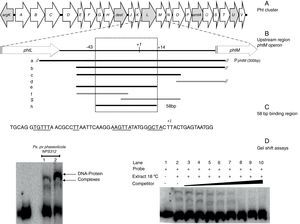
Gel shift assays. (A) Graphic representation of the Pht region. Each arrow represents an individual gene, with the direction of the arrow indicating the direction of transcription. Gray arrows indicate genes whose function have been previously reported. (B) Detailed view of the phtM operon upstream region indicating the fragments used as unlabeled DNA competitors. The phtL–phtM intergenic region consists of 149bp. The black bars represent the probes that were able to compete the DNA–protein complex, while the gray bars represent the probes unable to compete the complex. Fragment “h” corresponds to the 58 bp-region defined as binding site for protein. The truncated bars (//) are not represented to scale, whose length extends beyond what they are representing. (C) Nucleotide sequence of the 58bp binding region, showing the DNA regions common to promoters of phtD and phtM in bold type and underlined. (D) (Left) Gel shift assays evaluating binding capacity of the PphtM region using crude protein extracts of P. syringae pv. phaseolicola NPS3121 grown at 28°C (1) or 18°C (2) in M9 minimal medium; and (right) the gel shift competition assay performed using the “h” probe (58bp) as competitor of the labeled PphtM fragment. The concentration of unlabeled DNA competitors was as follows: lanes 1 and 2, no DNA competitor, lane 3, 25ng; lane 4, 50ng; lane 5, 60ng; lane 6, 100ng; lane 7, 150ng; lane 8, 200ng; lane 9, 250ng; lane 10, 300ng. The asterisks (*) indicate the DNA–protein complexes.
The probes used in gel shift assays (Table 1) were obtained by PCR amplification using the oligonucleotide pairs designed in this study. The double-stranded probes were end-labeled with (γ32P)-dATP using T4 polynucleotide kinase (Invitrogen). Gel shift assays were performed as previously described4. Briefly, protein extracts were prepared from P. syringae pv. phaseolicola NPS3121wt strain grown in M9 minimal medium at 18°C and 28°C until reaching the early stationary phase (OD600nm of 1.0). Cultures were centrifuged and the pellet was rinsed once with 1/20 volume of cold extraction buffer (25mM Tris–HCl pH 8.0, 0.1mM EDTA, 1mM DTT, 10% glycerol and 0.04mM PMSF), the cell pellet was freeze-thawed once, diluted in 1/20 volume of extraction buffer, and sonicated (3 times for 15s with intervals of 15s) in an ice bath using a Virsonic 60 sonicator (Virtis Company Inc). The cellular debris was pelleted by centrifugation at 15000×g in a microcentrifuge, for 5min at 4°C and discarded. Total protein was measured using the Bradford method with a BSA standard curve as control6. The binding reactions contained approximately 10ng of the probe, 30μg of the appropriate protein extract, 0.5–1μg poly(dI-dC) (Roche, Basel Switzerland), and 0.2μg sonicated salmon sperm DNA, in a 20μl total volume of binding buffer (25mM Tris pH 7.5, 50mM KCl, 1mM EDTA, 1mM DTT, 5% glycerol) and were incubated for 30min at room temperature. Protein-DNA complexes were separated from the unbound probe on 6.5% native polyacrylamide gels at 6mA for 3–4h, in 0.5× TBE buffer. Gels were vacuum-dried and exposed to a phosphor screen (Molecular Dynamics). The image was captured by scanning on a STORM 860 (Molecular Dynamics) and analyzed with Quantity One software (BIO-RAD, Berkeley, CA). To determine the specificity of the DNA-protein complexes observed, competition assays were carried out using increasing concentrations of specific and non-specific competitor DNA. A 300 bp-PvuII fragment of the pUC19 plasmid was used as non-specific competitor. To delimit the binding site of the putative regulatory protein and/or to determine the localization of the DNA–protein complex, competition assays were performed with an excess of unlabeled wild-type probes, listed in Table 1. When crude extracts of P. syringae pv. phaseolicola NPS3121 gacA− mutant, P. syringae pv. tomato DC3000 and P. syringae pv. phaseolicola CLY233 strains were assayed, the conditions of the gel shift assays were similar to those described above. The gel shift assays evaluating the upstream region of the phtD operon were performed using the PphtD probe previously reported under the conditions described above4.
Probes used in gel mobility shift assays
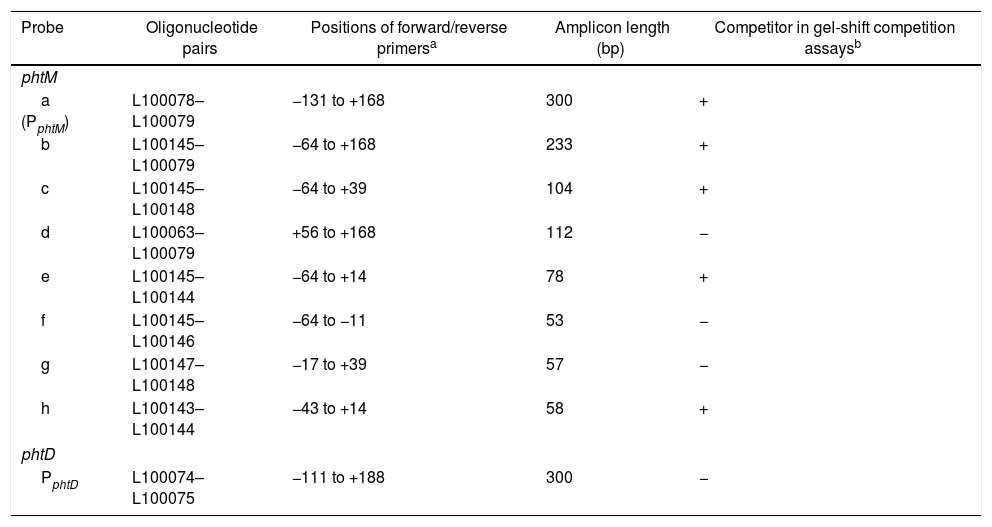
| Probe | Oligonucleotide pairs | Positions of forward/reverse primersa | Amplicon length (bp) | Competitor in gel-shift competition assaysb |
|---|---|---|---|---|
| phtM | ||||
| a (PphtM) | L100078–L100079 | −131 to +168 | 300 | + |
| b | L100145–L100079 | −64 to +168 | 233 | + |
| c | L100145–L100148 | −64 to +39 | 104 | + |
| d | L100063–L100079 | +56 to +168 | 112 | − |
| e | L100145–L100144 | −64 to +14 | 78 | + |
| f | L100145–L100146 | −64 to −11 | 53 | − |
| g | L100147–L100148 | −17 to +39 | 57 | − |
| h | L100143–L100144 | −43 to +14 | 58 | + |
| phtD | ||||
| PphtD | L100074–L100075 | −111 to +188 | 300 | − |
The Southwestern assays were performed as previously described with some modifications19. Briefly, protein extracts (70μg) prepared from P. syringae strains, under similar conditions to those used on the gel shift assays, were fractionated on a 12% sodium dodecyl sulfate (SDS) denaturing gel. The gels were transferred to a 0.45μm pore size nitrocellulose membrane by electroblotting under the following conditions: 10min at 30V, 10min at 40V and 40min at 50V. Blots were incubated with shaking in buffer TNE-50 (10mM Tris pH7.5, 50mM NaCl, 1mM EDTA, 1mM DTT) for 5min and subsequently incubated with shaking in blocking buffer SW (25mM HEPES pH 8.0, 1mM DTT, 10% glycerol, 50mM NaCl, 1mM EDTA, 2.5% low-fast skim milk) at 4°C for overnight. The blot was processed by washing one time in 50ml of TNE-50 buffer and incubated in hybridization buffer consisting in TNE-50 buffer, labeled target DNA (200ng) with 0.5–1μg of nonspecific competitor poly(dI-dC) DNA (Roche) at room temperature for 8h. The blot was processed by washing two times in 50ml of TNE-50 buffer and exposed to a phosphor screen (Molecular Dynamics). The image was captured by scanning on a STORM 860 (Molecular Dynamics) and analyzed with Quantity One software (BIO-RAD). Pre-stained protein markers were included in the assays for molecular mass estimation in Southwestern blots. To determine the specificity of the DNA–protein complexes observed, BSA protein was electrophoresed simultaneously. Additionally, competition assays were carried out using specific and non-specific competitor DNA. A 300 bp-Pvu II fragment of the pUC19 plasmid was used as non-specific competitor at 1μg of concentration. For the competition assays, the protein extracts were electrophoresed in duplicate in the same gel. After completion of the blocking, the blot membrane was divided into two parts; one was used as control employing the conditions above mentioned, while the other was used for the competition assays, in which the unlabeled competitor was added to TNE-50 hybridization buffer. To determine the ability to compete the DNA–protein complexes, the images of both blots were fused using Quantity-one software (BIO-RAD).
SDS-PAGE fractionationMolecular mass (MM) determination of unknown proteins by SDS-PAGE was performed as described previously26,36. Briefly, crude extracts (80μg) prepared from P. syringae pv. phaseolicola NPS3121wt strain grown in M9 minimal medium at 18°C, were electrophoresed on a 12% SDS-polyacrylamide gel under similar conditions to those mentioned above. The lane containing protein extracts was sliced uniformly into molecular mass intervals. Gel slices were crushed into 1.5 volumes of renaturation buffer (3% Triton X-100, 20mM Hepes, 100mM NaCl, 5mg/ml BSA, 3mM ZnCl2, 3mM MgCl2, 2mM DTT, 0.1mM PMSF, 0.1mM benzamidine–HCl) and incubated overnight at 4°C. The polyacrylamide debris was discarded by centrifugation, and the supernatant was then used to evaluate the DNA binding activity by gel shift assays using the reaction conditions previously mentioned. The PphtM probe was used in the gel shift assays. Prestained protein markers were included in the assays for molecular mass estimation. The proteins within the molecular mass intervals of 14–20kDa and 30–45kDa were analyzed by this technique.
ResultsThe promoter region of the phtM operon contains binding sites for putative regulatory proteinsIn order to identify potential transcriptional regulators of the phtM operon, we began this study by conducting an EMSA, to evaluate the binding activity of the phtM promoter region to proteins present in the extracts from P. syringae pv. phaseolicola NPS3121 grown at 18°C (the optimal temperature for toxin production) or 28°C. A 300bp radiolabeled DNA fragment (PphtM), spanning positions −131 to +167 relative to the transcription start site of the phM operon1 was used as probe (Fig. 1B). Mobility shift assays showed the presence of two specific DNA–protein complexes with extracts of cells grown at 18°C and 28°C (Fig. 1D) suggesting that the phtM promoter region contains a binding site for a putative regulatory protein whose presence, on the basis of the gel shift assays, is independent of temperature. Furthermore, the formation of two DNA–protein complexes also suggests the possible binding of two regulatory proteins and/or multimer protein on the phtM promoter region.
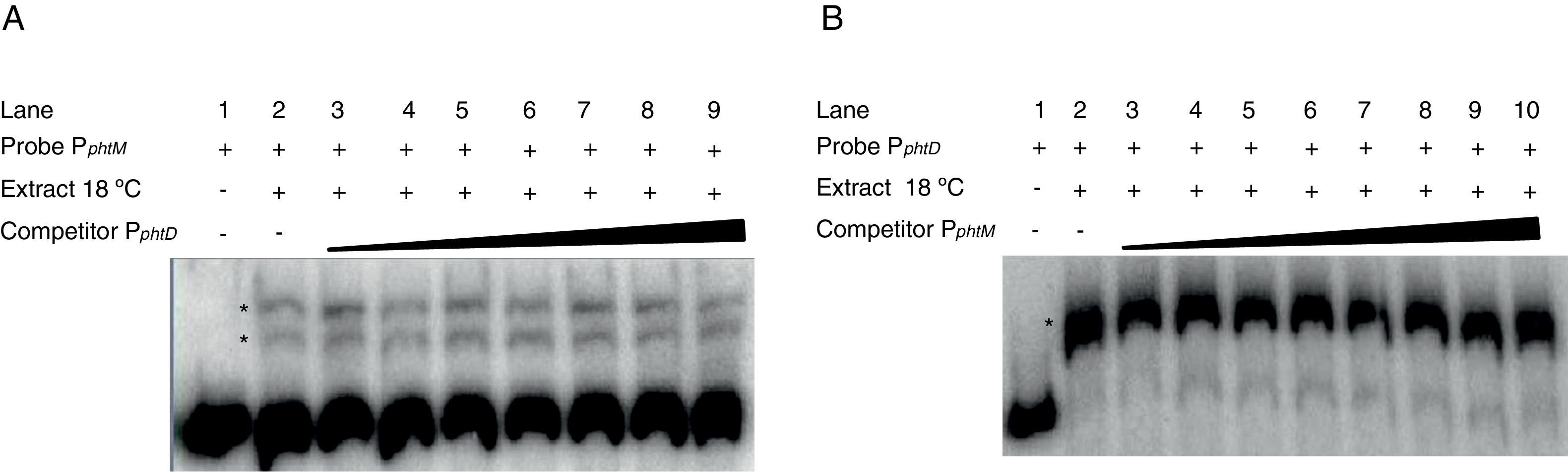
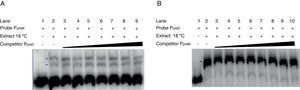
The phtM and phtD operons do not share a common mechanism of regulationPrevious works had suggested a common mechanism of transcriptional regulation for phtM and phtD operons due to the presence of conserved sequences in the promoter regions of both genes, which could be recognized by a similar or unique transcriptional regulator1. In order to evaluate whether the phtM and phtD promoters regions (PphtM and PphtD respectively) are targets of the putative regulatory protein, revealed through the gel shift assays, gel-shift competition assays were performed using the promoter region of the phtD operon (PphtD)4, as competitor into gel shift assays that evaluate the binding activity of the phtM promoter region (PphtM) and vice versa. The PphtM and PphtD regions contain the conserved sequences previously mentioned1. The results of these assays showed that the addition of the PphtD competitor in increasing concentrations does not interfere with the formation of protein–PphtM complexes, indicating that this PphtD site is not target of the putative regulatory protein of the phtM operon (Fig. 2A). Likewise, similar results were obtained when the PphtM region was used as competitor into the gel shift assays that evaluate the binding activity of the phtD promoter region (PphtD), indicating that PphtM is not a target of the phtD promoter binding protein, which has been previously identified as the IHF protein4 (Fig. 2B). These results suggest that these proteins, as revealed through the gel shift assays, are specific for the regulation of each operon.
Gel shift competition assays. (A) Gel shift competition assays using the PphtD region as competitor of the PphtM fragment. The concentrations of unlabeled DNA competitor used were: lanes 1 and 2, no DNA competitor; lane 3, 50ng (0.26pmol); lane 4, 60ng (0.30pmol); lane 5, 100ng (0.51pmol); lane 6, 150ng (0.77pmol); lane 7, 200ng (1pmol); lane 8, 250ng (1.3pmol) and lane 9, 300ng (1.5pmol). (B) Gel shift competition assays evaluating the competing capacity of the PphtM fragment on the PphtD–IHF interaction. The competitor was used in increasing concentrations: 50ng (0.26pmol); 60ng (0.30pmol); 100ng (0.51pmol); 150ng (0.77pmol); 200ng (1pmol); 250ng (1.3pmol) and 300ng (1.5pmol). The asterisks (*) indicate the DNA–protein complexes.
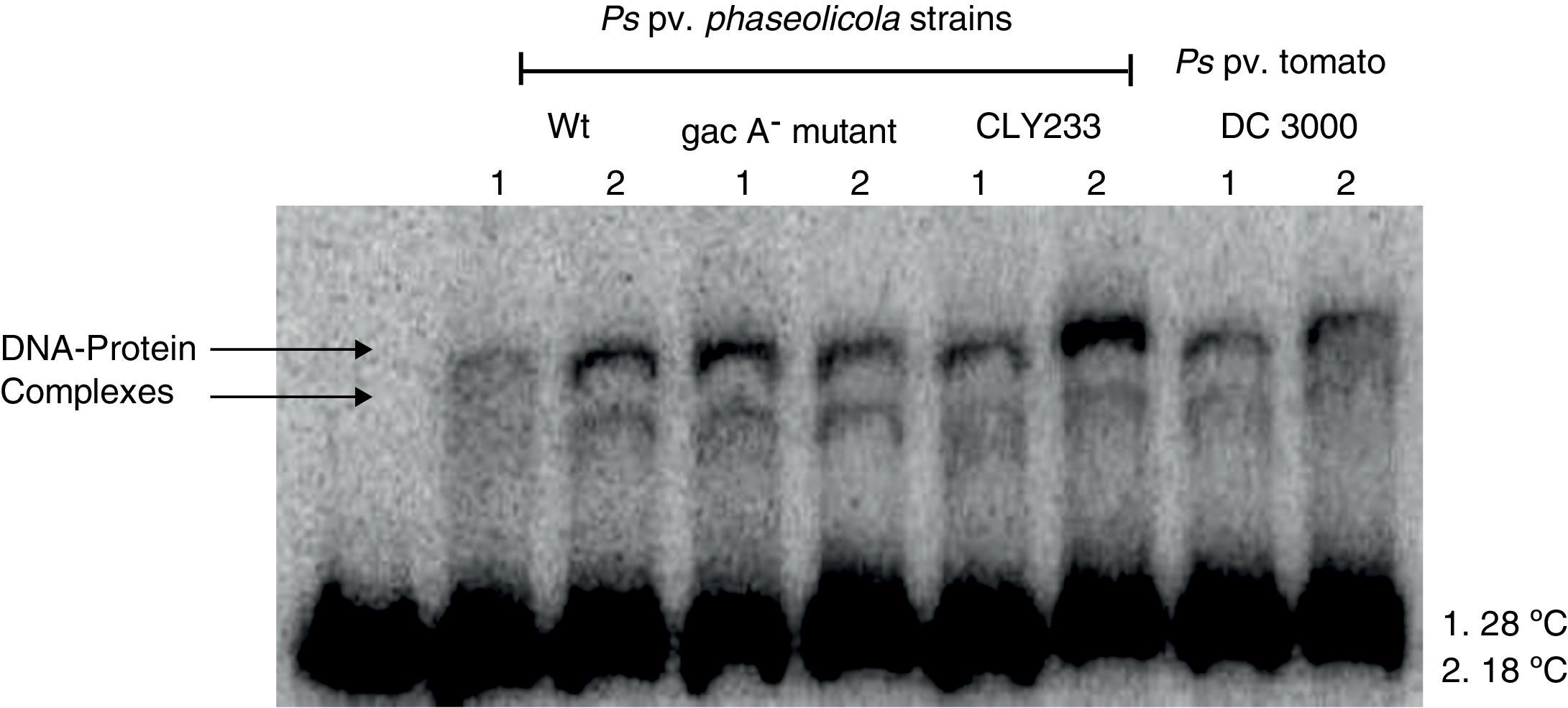
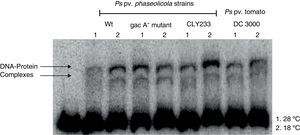
Since previous studies suggest that genes within the Pht cluster, particularly the phtL gene, could be involved in the regulation of the phtM operon1, we decided to evaluate whether the putative transcription factors of the phtM operon as revealed through the mobility shift analysis were encoded outside or within the Pht cluster. Gel-shift assays using crude extracts from P. syringae pv. phaseolicola strain CLY233, a non-toxigenic strain lacking the Pht cluster and P. syringae pv. tomato DC3000 (non-phaseolotoxin producer) grown at 18°C and 28°C in M9 minimal medium were performed. The incubation of the PphtM fragment with the protein extracts previously mentioned showed the presence of two identical DNA–protein complexes to those obtained with protein extracts from P. syringae pv. phaseolicola NPS3121 (Fig. 3). Gel-shift competition assays with specific and non-specific probes indicated that the observed DNA–protein binding was specific for the PphtM region (data not shown). These results indicate that the putative transcription factor binding upstream phtM is encoded by a gene located outside the Pht region and this is not specific for the phaseolotoxin synthesis due to the presence of this protein in other nonphaseolotoxin-producing P. syringae pathovars.
Gel shift assays with crude extracts of different pathovars of P. syringae. Radiolabeled PphtM fragment was incubated with protein extracts of P. syringae strains:pv. phaseolicola NPS3121wt and gacA− mutant; pv. phaseolicola CLY233 and pv. tomato DC3000, grown at 28°C (1) and 18°C (2) in M9 minimal medium.
Furthermore, because previous reports have suggested the influence of the GacS–GacA two component system on the expression of the Pht cluster genes by its effect on regulatory proteins9, we decided to evaluate whether the putative transcription factors binding upstream phtM are part of the hierarchical regulatory network of the GacS/GacA system. Gel-shift assays were performed using protein extracts from P. syringae pv. phaseolicola gacA− mutant grown at 18°C and 28°C in M9 minimal medium. Mobility shift assays showed the formation of two specific DNA–protein complexes in a similar position to that obtained with the wild type strain (Fig. 3). These assays suggest that the presence of the putative transcription factors binding upstream phtM is independent of the regulatory pathway of the GacS–GacA two-component system.
The binding site for the putative transcription factors of the phtM operon is located within a 58bp regionIn order to characterize the cis-acting elements involved in the binding of the putative regulatory proteins of the phtM operon, we performed gel shift competition assays to delimit the binding site of these proteins. Different fragments of the PphtM (Fig. 1B) region were used as unlabeled competitors in increasing concentrations. The results of these assays showed that the retarded band was effectively competed by fragments b, c, e and h (Table 1), thus indicating that the binding site of the regulatory protein is located in a 58bp region that spans positions −43 to +14, relative to the phtM operon transcription start site (Fig. 1C). Although fragments such as f and g, which span shorter regions within the h fragment (Fig. 1B, Table 1), were used in the competition assays, these were unable to compete the DNA–protein binding (supplementary material II). To evaluate the presence of conserved sequences to the 58bp region within the genome of the bacterium as well as in related P. syringae strains (P. syringae pv. tomato DC3000 and P. syringae pv. syringae B728), BLAST analyses were performed using the microbial nucleotide database optimizing for megablast, discontiguous megablast and BLASTn algorithms using default parameters. The results of these analyses showed that the “h” sequence does not exhibit similarity with any other region of the chromosome. These analyses showed that the 58bp region matched itself, with 100% identity in the regions of the available sequences of P. syringae pv. phaseolicola, such as the complete genome of the1448A strain (GenBank accession number CP000058.1); phaseolotoxin synthesis gene cluster of NPS3121 strain (DQ141263.1) and argK-tox gene cluster of pathovar phaseolicola (AB237164.1). Furthermore, the results showed that no similarity of the 58bp region was found with sequences of related P. syringae strains, with the exception of phaseolotoxin producer bacterium P. syringae pv. actinidae ICMP 9853 (CP018202.1) (data not shown).
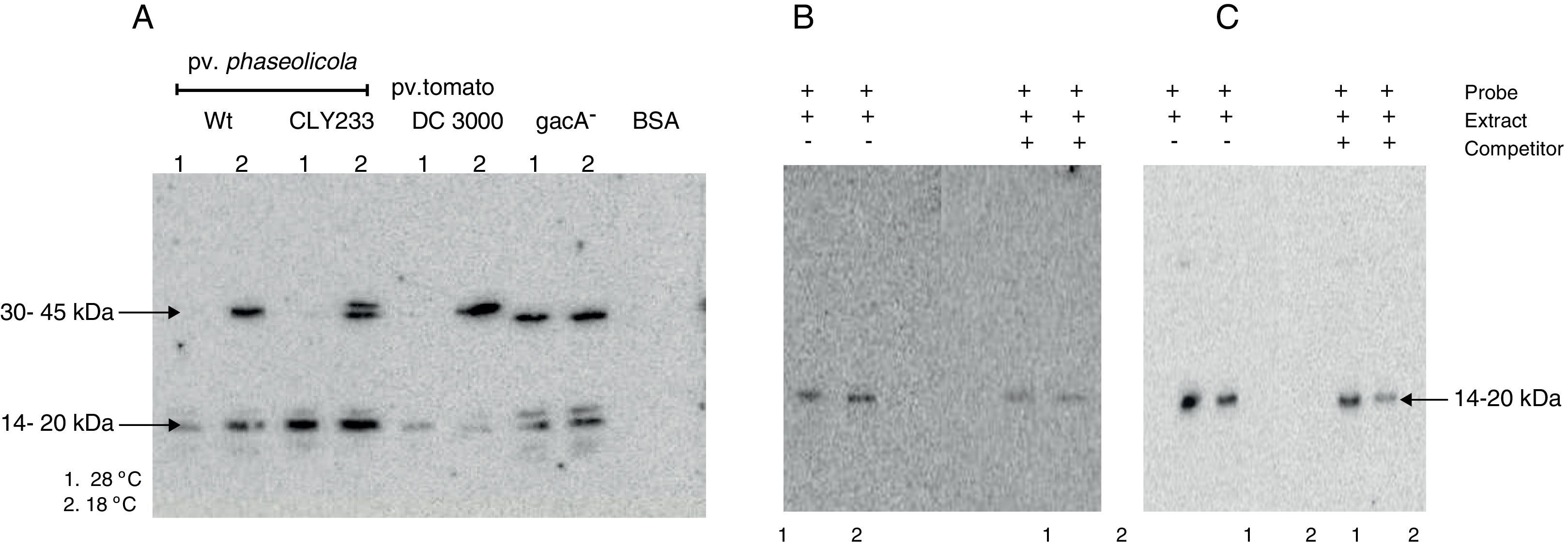
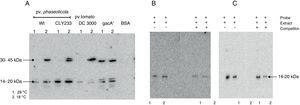
The putative transcription factor of the phtM operon has an apparent molecular mass in the 14–20kDa rangeOnce the binding site for the phtM regulatory protein had been delimited to a 58bp region, we evaluated the presence of putative cis-acting elements within the phtM promoter region using a transcription factor search program (BPROM, http://www.softberry.com). However the in silico analysis did not yield any positive results regarding transcription factors able to recognize this region. On the other hand, the search of inverted or direct repeats within the 58bp region by REPuter software18 using standard parameters showed the presence of three inverted repeats and two palindromic sequences with variable position and length within this region (data not shown). Therefore, in order to characterize the putative transcription factors binding upstream of the phtM operon, we performed Southwestern assays to estimate the molecular mass of these proteins. Protein extracts from P. syringae pv. phaseolicola NPS3121 strains wt and gacA− mutant, P. syringae pv. phaseolicola CLY233 and from P. syringae pv. tomato DC3000 grown at 18°C and 28°C, which contain the putative transcription factors, were electrophoresed on 12% SDS-polyacrylamide gels and incubated with the PphtM fragment. BSA protein was electrophoresed simultaneously and used as control to validate results. The assays identified a clear DNA binding activity with an apparent molecular mass in the 14–20kDa range in all evaluated cell extracts. Likewise, a DNA binding activity was identified with an apparent molecular mass in the 30–45kDa range solely with the protein extracts from P. syringae pathovars grown at 18°C with the exception of the gacA−mutant, in which the DNA binding activity was observed at both temperatures (Fig. 4A). Competition assays with specific probes indicated that the DNA binding activity observed in the 30–45kDa range appears to be unspecific because only those in the 14–20kDa range decrease in the presence of increasing concentrations of competitor (data not shown).
Southwestern assays and competition assays. (A) Southwestern assays with crude extracts of different pathovars of P. syringae grown at 28°C (1) or 18°C (2) using the PphtM fragment as probe. The arrows indicate the DNA–protein complexes. (B) Southwestern assay and competition assay using the “c” fragment of the upstream region of the phtM operon as probe (left) and as competitor (right). Fragment “c” contains the binding site for the putative regulatory protein. (C) Southwestern assay and competition assay using the “h” fragment of the phtM operon upstream region as probe (left) and as competitor (right). Fragment “h” corresponding to the 58 bp-region defined as binding site for the protein. 1=extract of P. syringae pv. phaseolicola NPS3121wt grown at 28°C, and 2=extract of P. syringae pv. phaseolicola NPS3121wt grown at 18°C.
To validate the above results, two types of additional experiments were performed: 1) Southwestern assays using the shorter fragments (“c” and “h”) as probes of the phtM promoter, which contain the binding site for the putative regulatory proteins and, 2) Molecular Mass (MM) Fractionation assays. The Southwestern assays were performed using protein extracts from P. syringae pv. phaseolicola NPS3121wt strain grown at 18°C and 28°C, which were electrophoresed on 12% SDS-polyacrylamide gels and incubated with the “c” fragment (104bp) of the phtM promoter region. The results of these assays showed only the presence of a polypeptide with DNA binding activity with molecular mass in the 14–20kDa range (Fig. 4B). Likewise, the Southwestern assays performed using the “h” fragment (58bp) as probe, corresponding to the minimal binding region of phtM regulatory proteins, identified a similar DNA binding activity in the 14–20kDa range (Fig. 4C). The competition assays showed that the DNA–protein binding observed is specific to the region (Fig. 4B, C). These results indicate that a putative regulatory protein of the phtM operon has an apparent molecular mass in the 14–20kDa range. However, given the constraints of the Southwestern assays using proteins under denaturing conditions, it is possible that the phtM protein may bind this region either as a monomer or a multimer of a single polypeptide.
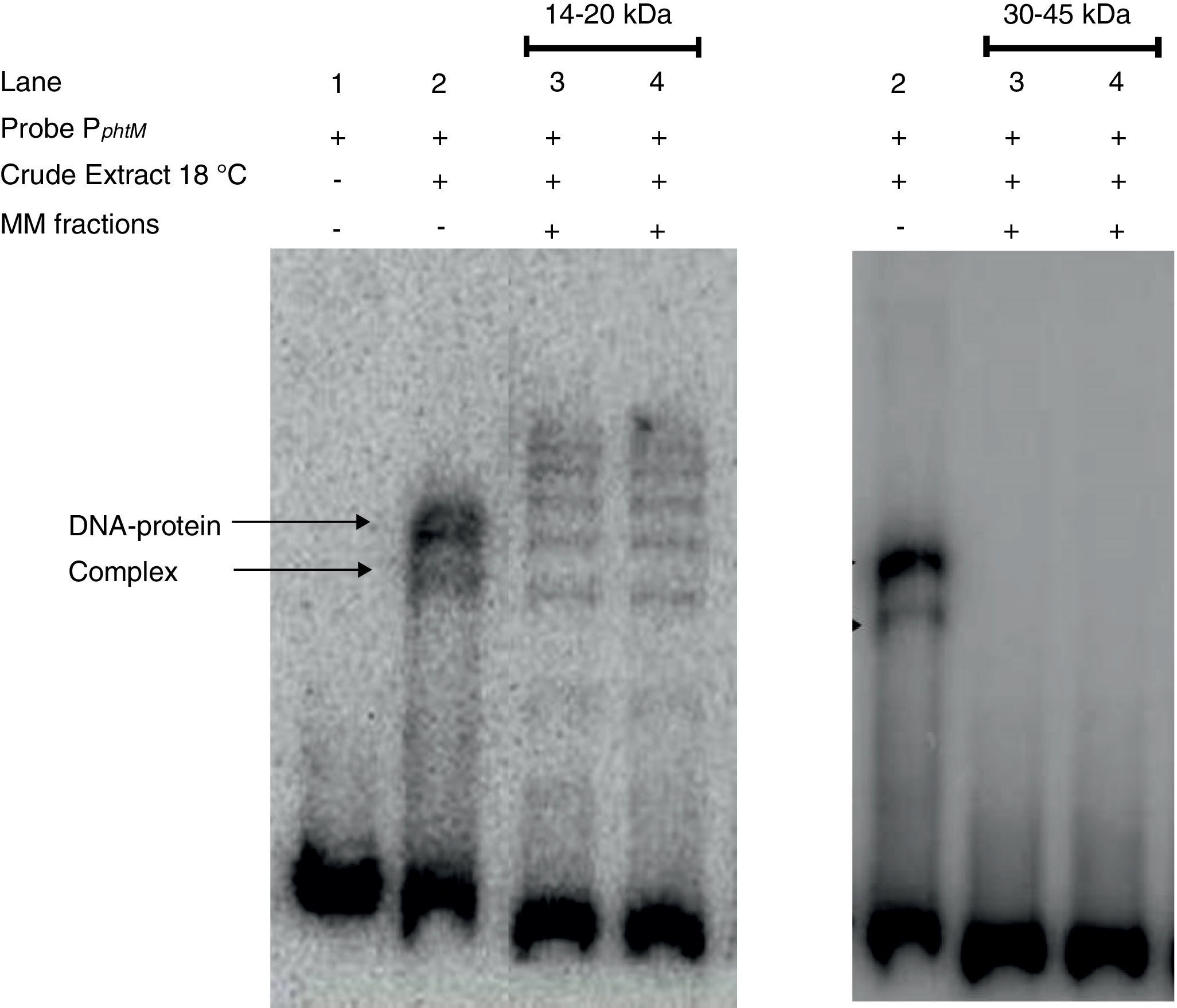
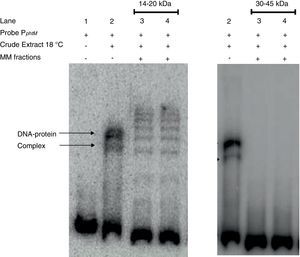
Finally, strong evidence concerning the molecular mass of the PphtM binding protein was obtained through SDS-PAGE-molecular mass (MM) fractionation. Protein extracts from P. syringae pv. phaseolicola NPS3121 grown at 18°C were electrophoresed on 12% SDS-polyacrylamide gels and fractions of molecular mass in 14–20kDa range and 30–45kDa range were excised and eluted from the gel. Proteins of each fraction were renatured and evaluated by gel shift assays for PphtM binding activity. The results showed that the incubation of the PphtM fragment (a) with proteins of molecular mass in the 14–20kDa interval were capable to produce retarding signals in identical position to those obtained with crude extracts from the wt strain, unlike proteins in the 30–45kDa range, which were unable to produce a DNA–protein complex (Fig. 5). Furthermore, the results of PphtM binding activity of the 14–20kDa fraction showed the formation of three other DNA–protein complexes that are not observed in the crude extract binding profile. These complexes might represent multimers of the phtM binding proteins. The results of these assays demonstrate that the phtM binding protein has a 14–20kDa molecular mass and it does not require additional proteins for its binding to the phtM promoter where it binds as monomer or multimer of a single polypeptide.
The phtM promoter binding protein corresponds to a 14–20kDa polypeptide. Protein extract of P. syringae pv. phaseolicola NPS3121 (wt) grown at 18°C was analyzed by SDS-PAGE, and Molecular Mass (MM) fractions in the 14–20kDa and 30–45kDa range were isolated in gel slices. Proteins were eluted, renatured and evaluated by EMSA. The PphtM–protein complex formation was detected in the 14–20kDa fraction. MM fractions were evaluated in duplicate.
Phaseolotoxin is an important virulence factor of P. syringae pv. phaseolicola and a key element for halo blight disease development in beans. The coding ability for the synthesis of this compound lies mainly in genes that can be found in the Pht cluster, and whose expression is mainly regulated by low temperature1. The knowledge about the regulatory pathways involved in the expression of the Pht cluster genes, and phaseolotoxin synthesis is very scarce, and studies focusing in this issue are necessary, to expand our understanding about the molecular basis governing these processes. In this work we initiated the search and characterization of transcription factors involved in the expression of genes of the phaseolotoxin synthesis (Pht cluster); particularly of the phtM operon in P. syringae pv. phaseolicola NPS3121.
The analysis of the upstream region of the phtM operon (PphtM) by electrophoretic mobility shift assays, clearly show the presence of two specific retarding signals, indicating that binding sites for regulatory proteins are found within this region. These results also indicated the possible binding of two regulatory proteins and/or multimer protein on the phtM promoter region, whose presence is independent of the temperature. Although the idea of a common mechanism of regulation for the phtD and phtM operons has been postulated, on the basis of the presence of six conserved sequences in the promoters of both genes1. The results of the gel competition assays using the phtM promoter region as competitor in the gel shift assays that evaluate the phtD promoter, clearly demonstrate that the upstream phtM region (PphtM) is not a target of the IHF protein, which has already been identified as a protein binding to the phtD promoter4. These results indicate that the identity of the putative regulatory proteins binding phtM, as revealed in the gel shift assays, does not correspond to an IHF protein. Furthermore, the results of the gel competition assays using the phtD promoter region as competitor in gel shift assays that evaluate the phtM promoter strongly suggest that specific regulatory pathways are involved in the expression of each transcriptional unit of the Pht cluster. However, we still cannot rule out the possibility that a common mechanism of regulation exists between the phtD and phtM operons, since the conserved sequences in these regions might be targets for a different protein, whose presence could not be observed under the conditions used in this study.
The assays with P. syringae pv. phaseolicola strain CLY233 (which lacks the Pht cluster) and P. syringae pv. tomato DC3000 (non phaseolotoxin-producer) show the formation of two DNA–protein complexes in identical position to that obtained with our working strain, indicating that the putative regulatory proteins of phtM are not encoded within the Pht cluster, thus ruling out the PhtL protein as responsible for the formation of the DNA–protein complexes observed. The regulation of the phtM operon by the PhtL protein has been suggested on the basis of a lack of expression of genes within the phtM operon in a phtL− mutant background1. However, a previous work has postulated that the regulatory role of the PhtL protein on the Pht cluster genes may be indirect through its effect on the Fur global regulator14. Additionally, the results of these assays indicate that phtM binding proteins are non-exclusive of the phaseolicola pathovar due to their presence in other non phaseolotoxin-producing pathovars of P. syringae, further suggesting that these could be global regulators. These results confirm the previous conclusions made by our working group, which indicate that the Pht cluster of genes has been integrated into the global regulatory mechanisms of the bacterium after the horizontal transfer event4. Some authors suggest that this event allows the host cells to control the expression of transferred genes thus avoiding unregulated expression that could have harmful consequences besides having a high energy cost10,20. The minimal region necessary for the binding of the putative regulatory proteins of phtM was delimited to a 58bp (−43 to +14) region, which still contains four of the six conserved sequences between the phtD and phtM promoters. Bioinformatics analysis showed that this region is not found in another part of the chromosome of the bacterium and further that it is not conserved in related P. syringae strains. Likewise, the bioinformatic analyses did not identify any known target sequences for transcription factors within this region, therefore preventing any possible inference regarding the regulatory pathways involved in the process. Nevertheless, previous research has found that the position of the binding site on the DNA relative to the transcription start site is indicative of the regulatory function of the protein. Thus, the repressors have downstream binding sites in relation to the RNA polymerase binding site at −35, while the large majority of activators have upstream binding sites5. This idea could suggest a negative role for the proteins binding upstream of the phtM region. However, because some atypical activators have demonstrated binding in downstream sites to the RNA polymerase binding site at −35, so far a positive regulatory function for the binding phtM protein5 could not be ruled out. More experimental evidence is necessary to determine its regulatory role. Experiments focused on the characterization of the binding protein to the phtM promoter may allow us to speculate about the action mode of the putative regulatory protein and about those characteristics or properties providing support to our ideas. In this sense, the Southwestern blot assays and SDS-PAGE-molecular mass (MM) fractionation using the a (PphtM), c and h probes of the phtM promoter show a clear DNA-binding activity in proteins present in the 14–20kDa range, indicating that the putative transcription factor of the phtM operon has a molecular mass within this range. Furthermore, on the basis of the constrained conditions of these assays, the results demonstrate that the putative transcription factor recognizes and binds to the upstream phtM region as monomer o multimer of a single polypeptide. Experimental work is still necessary to determine the identity of the regulatory protein of the phtM operon. In conclusion, the results of this study have allowed us to gain insight about the regulatory pathways involved in the phaseolotoxin synthesis and to further lay the groundwork for future research. From the data obtained in this study it is possible to device strategies aimed to the purification and identification of this putative regulatory protein of the phtM operon eventually leading to the identification of the gene/operon that encodes this protein.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestAll authors declare no competing interests.