Laguna Azul is an oligotrophic lake situated at 4,560m above sea level and subject to a high level of solar radiation. Bacterioplankton community composition (BCC) was analysed by denaturing gradient gel electrophoresis and the impact of solar ultraviolet radiation was assessed by measuring cyclobutane pyrimidine dimers (CPD). Furthermore, pure cultures of Acinetobacter johnsonii A2 and Rhodococcus sp. A5 were exposed simulta-neously and CPD accumulation was studied. Gel analyses generated a total of 7 sequences belonging to Alpha-proteobacteria (1 band), Beta-proteobacteria (1 band), Bacteroidetes (2 bands), Actinobacteria (1 band), and Firmicutes (1 band). DGGE profiles showed minimal changes in BCC and no CPD was detected even though a high level of damage was found in biodosimeters. A. johnsonii A2 showed low level of DNA damage while Rhodococcus sp. A5 exhibited high resistance since no CPD were detected under natural UV-B exposure, suggesting that the bacterial community is well adapted to this highly solar irradiated environment.
La Laguna Azul es un ambiente oligotrófico localizado a 4560m de altura y sometido a elevados niveles de radiación solar. La composición de su comunidad bacterioplanctónica fue analizada empleando la técnica de electroforesis en gradiente desnaturalizante y se investigó el impacto de la radiación ultravioleta cuantificando los dímeros de pirimidina (CPD). Además, se expusieron simultáneamente cultivos puros de Acinetobacter johnsonii A2 y Rhodococcus sp. A5 para estudiar la acumulación de CPD. El análisis de los geles mostró siete secuencias pertenecientes a Alpha-proteobacteria (1 banda), Beta-proteobacteria (1 banda), Bacteroidetes (2 bandas), Actinobacteria (1 banda) y Firmicutes (1 banda). A lo largo del día se observaron cambios mínimos en la composición de la comunidad y no se detectaron CPD. A. johnsonii A2 presentó un daño bajo mientras que Rhodococcus sp. A5 no presentó daño en su ADN, sugiriendo que la comunidad bacteriana está muy bien adaptada a este ambiente altamente irradiado.
High Altitude Andean Wetlands (HAAW) have a broad range of extreme conditions such as high solar radiation fluxes due to the combination of atmospheric and geographical factors1,4. Among the environmental factors, solar radiation and particularly ultraviolet radiation (UVR, 280–400nm) may have strong effects on the production, activity and abundance of bacterioplankton13. In general, UVR causes DNA damage. Ultraviolet B radiation (UV-B, 280–320nm) causes direct damage through the generation of cyclobutane pyrimidine dimers (CPD). These lesions block DNA and RNA polymerase activities inhibiting gene replication and transcription8.
Previous studies at our lab demonstrated that the bacterial communities from Laguna Negra and Laguna Verde, both located next to Laguna Azul, were well adapted to long-term artificial UV-B exposure, and in many cases UV-B radiation even stimulated growth11. In situ exposure to natural radiation of water samples from Laguna Vilama, a hyper saline lake located in the Argentinean Puna at 4,660m above sea level (a.s.l.), suggest that the bacterioplankton community is well adapted to this highly solar irradiated environment showing little accumulation of CPD and few changes in the community composition9.
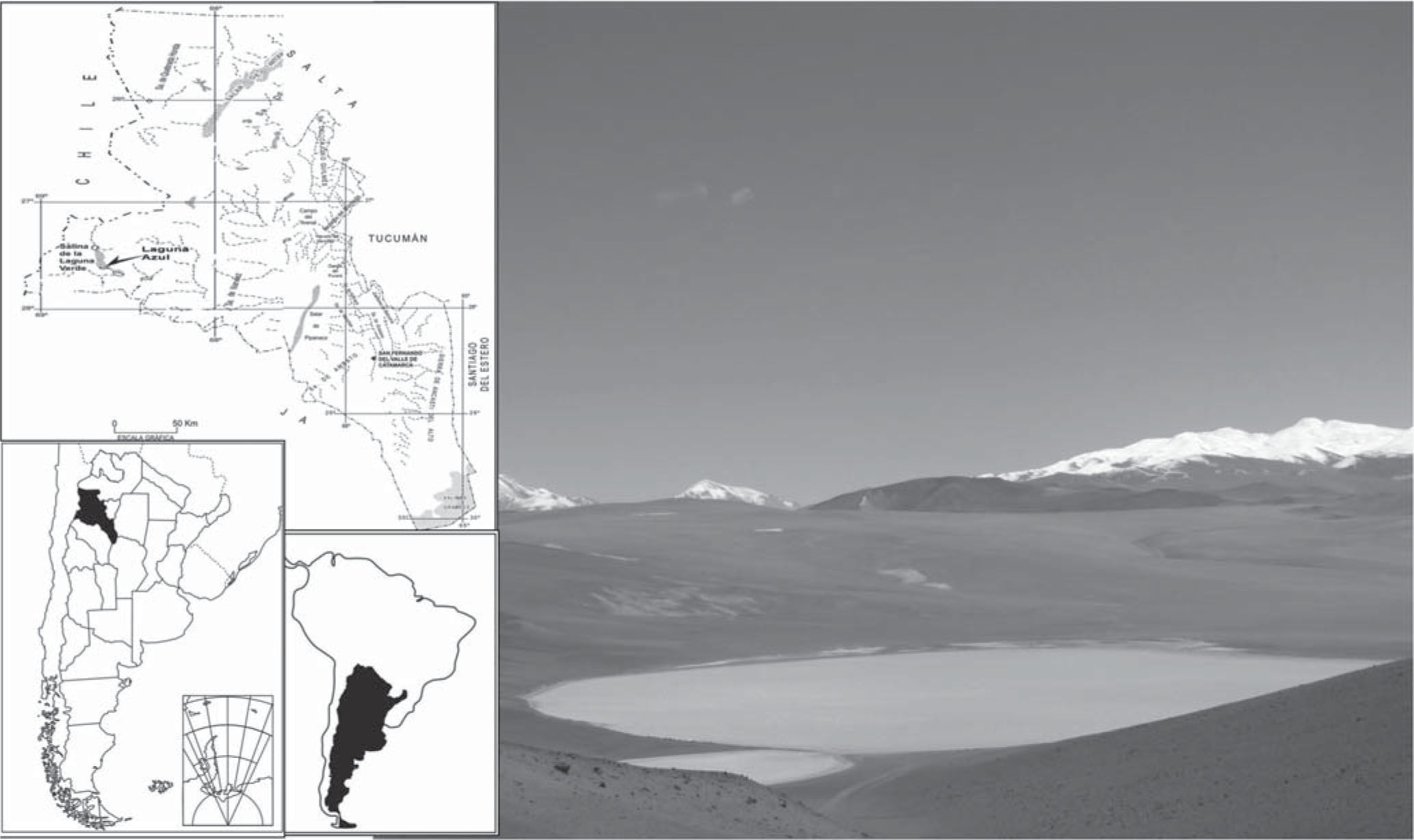
In this paper we carried out a survey of bacterioplankton diversity in Laguna Azul (27° 34’ S and 65.8° 32’ W; 4,560m a.s.l.) (Fig. 1) by using a fingerprinting technique (DGGE) and we assessed DNA damage in the plankton community, pure cultures of Acinetobacter johnsonii A2 and Rhodococcus sp. A5 and biodosimeters measuring CPD throughout one day. Incident solar radiation incident over the studied area was monitored during November 2006 (austral spring) using a broad-band European Light Dosimeter Network (ELDONET) radiometer (Real Time Computers). On the sampling day, the maximum UV-B (280–315nm) irradiance reached 3.3 Wm−2 at 11:30 (local hour), UV-A (315–400nm) irradiance was 25.6 Wm−2 and PAR (400–700nm) was 340 Wm−2 (data no shown).
Surface water samples were collected with a clean bucket to be used for experiments. Water was pre-filtered through a 100μm Nitex mesh and duplicate samples were placed in 800ml Plexiglas bottles that received full radiation (UVR+PAR, 280–700nm). The containers were then placed into 100μm Nitex mesh frames that were attached to a buoy. At the beginning of the experiment, sub-samples were processed for the determination of bacterioplankton composition by DGGE and CPD. Incubation was started at 11:00 (local hour) and 50ml aliquots were collected, by duplicate, at 13:00 and 15:00. Then, water samples were filtered through 0.22μm polycarbonate filters, immediately frozen in liquid nitrogen and kept at −20°C until DNA extraction. In addition, pure cultures of A. johnsonii A2 and Rhodococcus sp. A5, two bacteria previously isolated from water of this lake6,15, were exposed to natural radiation simultaneously with the lake water. Bacterial suspensions were prepared in the laboratory as it was described by Farias et al.9 and maintained at 4°C in the dark until in situ exposure. In Laguna Azul, the Plexiglas bottles containing bacterial suspension were placed in anodized aluminum frames. After the incubation period, the samples were processed to determine CPD accumulation. Controls of unexposed samples were run simultaneously in darkness by wrapping the Plexiglas bottles with aluminum foil.
DNA biodosimeters (naked calf thymus DNA used as control of DNA damage) were also incubated in situ to determine the DNA effective dose3. The CTAB protocol of DNA extraction was used10. To determine changes in bacterial assemblages, the variable V3 region of 16S was enzymatically amplified by PCR with primers to conserved regions of the 16S rRNA genes. The nucleotide sequences of the primers are as follows: primer 1 F341-GC: 5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC GCC TAC GGG AGG CAG CAG-3′, primer 2 R518: 5′-CGT ATT ACC GCG GCT GCT GG-3′ and primer 3 F357: 5′-TAC TGA TAG AA T GTG GAG C-3′.
PCR amplification and DGGE were performed as it was described previously by Farias et al.9 Distinguishable bands were excised from the gel; the eluted DNA was used as template for re-amplification using primers 2 and 3 and PCR products were sequenced in Macrogen Korea.
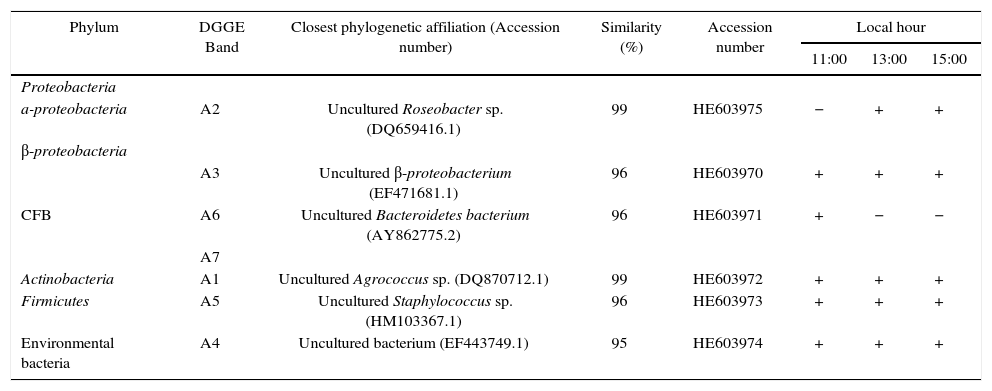
Gel analysis gave a total of 15 predominant bands, defined by intensity and frequency of appearance. The 16S rRNA gene sequence of DGGE bands obtained at all sampling times belonged to Alpha and Beta-proteobacteria (Roseobacter sp. and uncultured Beta-proteobacterium), CFB (an uncultured Bacteroidetes), HGC (Agrococcus sp.) and LGC (Staphylococcus sp.). Minor modifications in the community structure were observed throughout the day. Uncultured Beta-proteobacteria (band A3), Agrococcus sp. (band A1) and uncultured bacteria (band A4) were present at all sampling times. Bands that disappeared with increasing solar radiation doses were related to Bacteroidetes (bands A6 and A7). Finally, a band that emerged during solar exposition was related to Roseobacter sp. (band A2) and Staphylococcus sp. (band A5) (Table 1).
Identification of DGGE bands from Laguna Azul and ch anges during one-day exposure
| Phylum | DGGE Band | Closest phylogenetic affiliation (Accession number) | Similarity (%) | Accession number | Local hour | ||
|---|---|---|---|---|---|---|---|
| 11:00 | 13:00 | 15:00 | |||||
| Proteobacteria | |||||||
| a-proteobacteria | A2 | Uncultured Roseobacter sp. (DQ659416.1) | 99 | HE603975 | − | + | + |
| β-proteobacteria | |||||||
| CFB | A3 | Uncultured β-proteobacterium (EF471681.1) | 96 | HE603970 | + | + | + |
| A6 | Uncultured Bacteroidetes bacterium (AY862775.2) | 96 | HE603971 | + | − | − | |
| A7 | |||||||
| Actinobacteria | A1 | Uncultured Agrococcus sp. (DQ870712.1) | 99 | HE603972 | + | + | + |
| Firmicutes | A5 | Uncultured Staphylococcus sp. (HM103367.1) | 96 | HE603973 | + | + | + |
| Environmental bacteria | A4 | Uncultured bacterium (EF443749.1) | 95 | HE603974 | + | + | + |
+ presence, − absence of band.
The amount of CPD was determined by using the method proposed by Boelen et al.3 employing a primary antibody (H3, Affitech, Oslo) directed mainly to thymine dimers. CPD detection was done using ECL detection reagents (Amersham) in combination with photosensitive films (Amersham Hyperfilm ECL). Films were scanned and analysed using an image analyser (Gel Doc 2000 Bio Rad). Each blot contained two dilution series of standard DNA with known amounts of CPD, allowing the estimation of absolute amounts of CPD.
No damage in the DNA was detected in the planktonic community during exposure to solar radiation. A. johnsonii A2 showed 21.6±2.55 CPD Mb−1 at the end of the exposure and no CPD were detected in Rhodococcus sp. A5 saline suspension exposure to natural radiation. However, 1,763±174.44 CPD Mb−1 were detected in the calf thymus DNA at the end of the experiments.
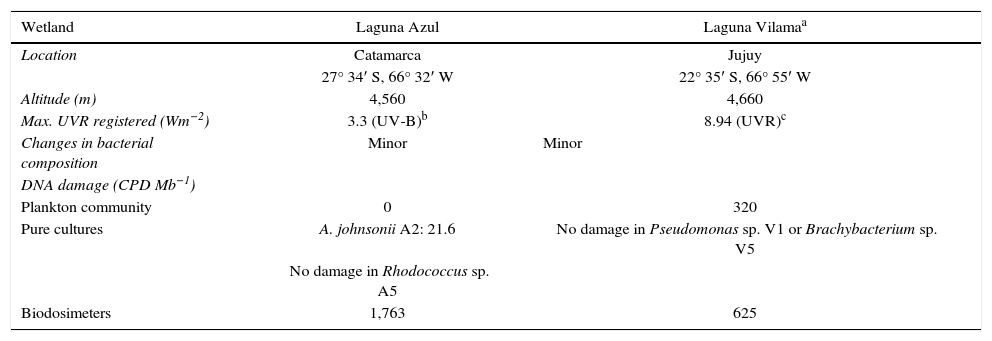
The present study contributes to our knowledge of the bacterial diversity of a remote and rather unexplored habitat and the effect of natural radiation in this microbial community. The microbial community in Laguna Azul was similar to those from other Argentinean and Chilean high-altitude wetlands where Bacteroidetes and Proteobacteria were predominant5,7,9,12. As it was observed in Laguna Vilama, a HAAW where similar experiments were carried out9, no significant changes in the bacterioplankton community composition throughout a day were detected (Table 2). The bands recovered in Laguna Azul revealed sequences shared with Laguna Vilama such as Agrococcus sp. and Roseobacter sp. In both lakes, the uncultured Agrococcus band remained stable during solar exposure while in Laguna Azul, the Roseobacter sp. band increased its intensity during solar exposure.
Summary of similar parameters measured in Laguna Azul and Laguna Vilama
| Wetland | Laguna Azul | Laguna Vilamaa |
|---|---|---|
| Location | Catamarca | Jujuy |
| 27° 34′ S, 66° 32′ W | 22° 35′ S, 66° 55′ W | |
| Altitude (m) | 4,560 | 4,660 |
| Max. UVR registered (Wm−2) | 3.3 (UV-B)b | 8.94 (UVR)c |
| Changes in bacterial composition | Minor | Minor |
| DNA damage (CPD Mb−1) | ||
| Plankton community | 0 | 320 |
| Pure cultures | A. johnsonii A2: 21.6 | No damage in Pseudomonas sp. V1 or Brachybacterium sp. V5 |
| No damage in Rhodococcus sp. A5 | ||
| Biodosimeters | 1,763 | 625 |
With regard to the band related to Staphylococcus, an increase in its intensity was observed during natural solar exposure in Laguna Azul. In a previous work, this genus was described as UV-resistant since long-term exposure of water samples from this lake showed that a Staphylococcus-related isolate had high resistance to artificial UV-B radiation15. A similar response was observed in other HAAW where long-term water exposure to UV-B showed that a strain related to Staphylococcus was one of the most resistant in the assay conditions11.
Contrary to Laguna Vilama, no damage in the DNA in plankton from Laguna Azul was detected (Table 2). However the high amount of CPD detected in biodosimeters in Laguna Azul would indicate the presence of efficient protection systems in the planktonic community from this environment.
In early works6,15Rhodococcus sp. A5 strain was identified as Nocardia sp. A5 but deep physiological and genetic studies allowed to reassign it to the genus Rhodococcus and renamed it as Rhodococcus sp. A514. In agreement with previous works high resistance to artificial UV-B radiation was described2,14,15, both A. johnsonii A2 and Rhodococcus sp. A5 showed high UV-B resistance in their native environment since low or no damage were detected in their DNA.
In conclusion, DNA damage over the total microbial community as well as in individual strains from Laguna Azul showed that microorganisms living in such condition have developed efficient DNA protection systems to overcome irradiation stress. Currently, metagenomic studies are carried out in order to reveal the genetic mechanisms related to DNA repair.
Ethical disclosuresProtection of human and animal subjects. The authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of data. The authors declare that no patient data appear in this article.
Right to privacy and informed consent. The authors declare that no patient data appear in this article.
Conflicts of interestThe authors declare that they have no conflicts of interest.
We thank Dr. Julián Dib for his comments and corrections to the Ms. We also thank Regina Flores and Omar Ordoñez for technical assistance. This work was supported by PICT 1707 and 1788.