The driving forces behind many soil processes are microorganisms and they are able to respond immediately to environmental changes. The soil microbial community impacts on many soil properties. More than one-third of the terrestrial ecosystems are semiarid. However, a limited number of studies have been conducted to characterize soil fungal communities in semiarid grasslands, in particular those of agricultural fields. The aim of this study was to explore changes in the diversity and structure of soil fungal communities in semiarid grasslands, after different doses of glyphosate were applied under field conditions. Changes in soil fungal communities were examined using different approaches including culturing, calcofluor white stain and denaturing gradient gel electrophoresis (DGGE). The different approaches complement each other, revealing different aspects of the effect of glyphosate on soil fungal communities. We demonstrated a negative effect of glyphosate on soil fungal biomass at high doses and an early and transitory stimulatory effect on soil fungal biomass. We also found a negative effect of glyphosate on the species richness of cultivable fungi and changes in the molecular structure of soil fungal communities after double doses or long-term glyphosate application. In summary, our findings demonstrate an overall negative effect of glyphosate on soil fungal communities.
Los microorganismos del suelo son los responsables de llevar a cabo la mayoría de los procesos biológicos que ocurren en el suelo, y son capaces de reaccionar ante el estrés ambiental. Más de un tercio de los ecosistemas terrestres son semiáridos. Sin embargo, son escasos los estudios realizados para caracterizar las comunidades fúngicas en suelos agrícolas en ecosistemas semiáridos. El objetivo del presente trabajo fue estudiar los cambios que se producen en la biomasa, la diversidad y la estructura de las comunidades fúngicas del suelo, luego de la aplicación de distintas dosis de glifosato en condiciones de campo. Se emplearon diferentes técnicas incluidas el cultivo, la tinción directa con blanco de calcoflúor y PCR acoplada a electroforesis en geles de gradiente desnaturalizante (DGGE). Las distintas metodologías empleadas se complementan entre sí al detectar cada una distintos aspectos del efecto del glifosato en las comunidades fúngicas del suelo. Se encontró que el glifosato produce un efecto negativo sobre la biomasa fúngica, también se encontró un efecto transitorio estimulante inmediatamente posterior a la aplicación del herbicida. Además, se vio un efecto negativo sobre la riqueza de hongos cultivables, así como también cambios en la estructura molecular de las comunidades luego de aplicaciones repetidas. En conclusión, se demostró un efecto negativo generalizado sobre las comunidades fúngicas del suelo.
Agricultural practices, especially intensive ones, rely on pesticides for protecting crops and maximizing yields and economic benefits. Among the pesticides, herbicides are the most widely used. Nowadays, glyphosate is the most used and the most questioned herbicide51. The development and introduction of glyphosate-tolerant plant species could be explained by an exponential increment in the use of herbicides containing glyphosate in recent years. Since the mid-1990s, significant changes have occurred in when and how glyphosate herbicides are applied, and there has been a dramatic increase in the total volume applied7
Glyphosate [N-(phosphonomethyl) glycine] is a systemic, non-selective, post-emergence herbicide; it constitutes the active ingredient of more than 750 different broad-spectrum herbicides used worldwide. Recommended glyphosate field doses vary between 0.96 and 2.88kg a.i./Ha but high doses or repeated applications are common15,27,59. This herbicide has a moderate persistence in soil and is degraded predominantly by co-metabolic microbial processes40,41. Glyphosate targets a key enzyme in the shikimate pathway (5-enolpyruvyl-shikimate-3-phosphate synthase) involved in the production of aromatic amino acids (phenylalanine, tyrosine and tryptophan) that are essential for the growth of most plants, inhibiting nucleic acid metabolism and protein synthesis10. Moreover, the shikimate pathway is present in non-target organisms such as bacteria, fungi and algae.
Semiarid and arid lands cover more than one-third of the terrestrial ecosystems on Earth, and they are one of the most relevant ecological areas38,55. The expansion of no-till practices and the increase in genetically modified crops have resulted in a continuous increment of herbicide input17. The Argentinian Pampas is a wide plain with more than 52 million hectares of land suitable for cattle rearing and cropping55.
Agricultural practices and the deposition of agrochemical residues in soil induce changes in the microbial communities present in these environments40,41. Soil microorganisms are the driving force behind many soil processes including transformation of organic matter, nutrient release and xenobiotic degradation. Microorganisms are expected to be more sensitive and more efficient indicators than physical or chemical parameters as they are able to respond immediately to environmental changes23. The soil microbial community impacts on many soil properties, such as quality, plant health, long-term soil sustainability, resistance to perturbations, suppression of plant disease and even improvement of plant stress tolerance7. Examining the effects of glyphosate on soil microbial communities is relevant due to the critical role microorganisms play in soil processes, ultimately enabling ecosystems to function and provide services to humanity. In agricultural systems, the non-target effects that glyphosate could have on soil microbial communities are now of concern since they negatively affect soil functions, plant health, and crop productivity34. There are numerous reports on the harmful effects of glyphosate on microorganisms under laboratory conditions2,5,8,13,23,26,34 and extensive research has been done on the short-term response of soil microorganisms to glyphosate; however, relatively little information is available on the effects under field conditions11,12,41. The field experiments are more complex, but they are important to validate the studies performed in vitro at a laboratory scale. Even though controlled laboratory conditions allow for a detailed analysis of the effect of one or a few factors at a time while other factors are kept constant, these do not reflect the field conditions in which the fungi interact with the herbicide.
Fungi are the most diverse group of soil microorganisms and they are involved in almost all soil processes, playing an important role in the formation of stable aggregates and the maintenance of soil structure20. The relationship of glyphosate to fungi varies from toxicity to biodegradation. Glyphosate toxicity was demonstrated in vitro by Busse et al. (2001)12; however, when recommended field doses were tested in a soil microcosm assay, the changes in soil fungal communities were inconsistent41. Several studies have shown that glyphosate may exert at least temporary changes in soil microbial activity5,19, inhibiting the growth of some fungal species and stimulating others, including plant pathogens23,29. Several species in Aspergillus or Fusarium, are able to degrade glyphosate and could be recommended as bioremediation agents in contaminated soils37.
The analysis of ecological, structural and functional properties of soil microbial communities can allow a better understanding of the effect of pesticides on them, under field conditions21. The aim of this study was to investigate the effect that recommended and double doses of glyphosate had on the biomass and molecular profiles of soil fungal communities. We applied an integrated approach to evaluate the effect of glyphosate on soil fungi in a temperate agricultural Mollisol from a semiarid area.
Materials and methodsStudy area, experiment design and soil samplingThe study area is located in an experimental field belonging to the Universidad Nacional del Sur (38°29́56″ S; 62°17́36″ W). The climate is temperate and semiarid with little rainfall and a mean annual temperature of 15°C35. The soil type is classified as Mollisoll with 2% organic matter content9,35,49. Values of temperature (max, min, average) and rainfall were recorded throughout the experiment. The experiment was laid out in a randomized complete block design with two treatments and a control, with four blocks subdivided into three plots of 3.6m2 each. The herbicide employed was a commercial formulation of glyphosate (N-phosphonomethyl-glycine, 48% amine salt, Atanor Laboratories). The plots were treated with glyphosate 1× (0.96kg i.a/Ha), glyphosate 2× (1.92kg i.a/Ha), or 2l of water (control). Herbicides were prepared in situ and applied immediately. Samples were taken over two consecutive years, on a weekly schedule (2, 5–7, 13, 21–26 days after application). Due to weather conditions, the third week sample of the second year has missing data and an additional subsequent sample was taken (45 days after application). Five soil cores of 20g each were taken from the surface layer to a depth of 0–10cm in different parts of each plot and then mixed to make a composite sample. Samples were preserved at 4°C in the dark prior to processing. Then they were kept at −20°C for molecular analysis.
Fungal biomass estimation using a culture-dependent methodViable counts of fungi were performed in Rose Bengal-Chloramphenicol agar. Biomass estimates based on microbial counts were performed according to the following general expression Biomass=No. cells×volume x density, where the number of cells corresponded to the CFU/g dry soil. The density was 1.2g/cm and the hyphal volume was calculated considering a diameter of 5μm for fungal hyphae and 10m length of fungal mycelium per gram of soil. Individual colonies were transferred to sterile solid medium for morphological identification. All the isolated species were observed under a light microscope and only the colonies with reproductive structures were identified.
Culture independent approach: direct estimation of fungal biomassEstimation was performed using fluorescence microscopy and soil smears according to Vázquez et al.54 The hyphal length (μm/grid) was estimated by counting the number of intersections of hyphae with all the lines of the counting grid. Four smears per soil sample were prepared and counts were performed in five random fields per soil smear.
Culture independent approach: Denaturalization gradient gel electrophoresis (DGGE)
Extraction of DNA from soil samples: Soil samples were conditioned at 20°C for 24h. Forty composite samples were made by mixing the samples from the four blocks. Total microbial DNA was extracted from a 0.25g soil sample using the MoBioPowerSoil DNA Isolation Kit (MoBio Laboratories Inc. Carlsbad, CA, USA) as recommended by the manufacturer. The extracted DNAs were quantified using QubitTMfluorometer (Invitrogen) and checked by 0.7% (w/v) agarose gel electrophoresis with 3μl gel red (GenBiotech) in TBE 5× buffer (0.54g Tris, 0.275g boric acid, 0.2μl EDTA, pH=8) and visualized under UV light. The DNAs were then stored at −20°C.
Amplification of soil fungal DNA: Amplification of fungal 18S rDNA from soil DNA extracts was carried out using the primer pairs EF4 and ITS456 followed by re-amplification using the primer pairs ITS1F-GC16,33 and ITS256. PCRs were carried out on an XP Cycler (Bioer) in 25μl reaction volumes containing: 100ng of template DNA, 0.5μl each primer (50μM), 0.125μl (5mM) each dATP, dCTP, dGTP, dTTP, 2.5μl Cl2Mg (2.5mM), 2.5μl 10 x buffer, 13.3–17.3μl sterile distilled water (to complete the reaction volumes to 25μl) and 0.2μl TaqPolymerasa (1U). Cycling parameters were 95°C for 3min, followed by 25 cycles of 94°C for 1min, 44 or 50°C for 1min (EF4/ITS4 and ITF1F-GC/ITS2 primer pairs, respectively) and 72°C for 2min with a final extension at 72°C for 10min. Reactions were performed in duplicate, and negative controls (containing no DNA) were included in each PCR. Amplification products were checked by electrophoresis in 1.5% (w/v) agarose gels with 3μl red gel in TBE 5× buffer and visualized under UV light. Amplification products were then stored at −20°C.
Separation of PCR amplicons by denaturing gradient gel electrophoresis: Polyacrylamide gels were prepared with 10% [0.7M urea–4% (v/v) formamide] to 50% [3.5M urea–20% (v/v) formamide] vertical denaturing gradient using a gradient former and a peristaltic pump with a flow rate of 4ml/min.3 The buffer solution was TAE 1× and each lane of the gels was loaded with 10μl of re-amplification PCR products. The gels were run at 70V for 6h at 60°C using the VS20-DGGE (Cleaver Scientific Ltd.). After electrophoresis, gels were silver-stained as previously described by McCaig et al.28 and were scanned using an EPSON CX-5600 scanner. For the analysis of DGGE fingerprints, a binary matrix showing the presence (1) and absence (0) of DGGE bands was used to calculate Jaccard's similarity coefficient. Cluster analysis was conducted with the NTSYS 2.1 software44 using the unweighted pair group method (UPGMA).
Statistical analysisThe climatic variables were compared between the studied seasons by ANOVA. We performed a Generalized Linear Model with a Gamma distribution and a log link function to analyze the variation of fungal biomass (g/kg soil and μg C fungal/g soil, estimated by culture-dependent and direct methods, respectively), with respect to the treatment, days after herbicide application, temperature (T), precipitation (PP), and the year. Gamma distribution supports positive continuous variables, being in some cases a more realistic alternative than a normal distribution. We tested which probability distribution best fitted the data using Q–Q plots and the gamma distribution showed to be the most appropriate.
We tested the effect of the herbicide treatment for each day separately. The statistical analysis was performed using a blocked design and the relevance of the explanatory variables was analyzed using Likelihood Ratio Test55. Pearson's correlation coefficient was calculated between soil fungal biomass estimated with culture dependent and independent methods. All statistical analyses and graphics were performed using R42 and the packages MASS and ggplot26,57.
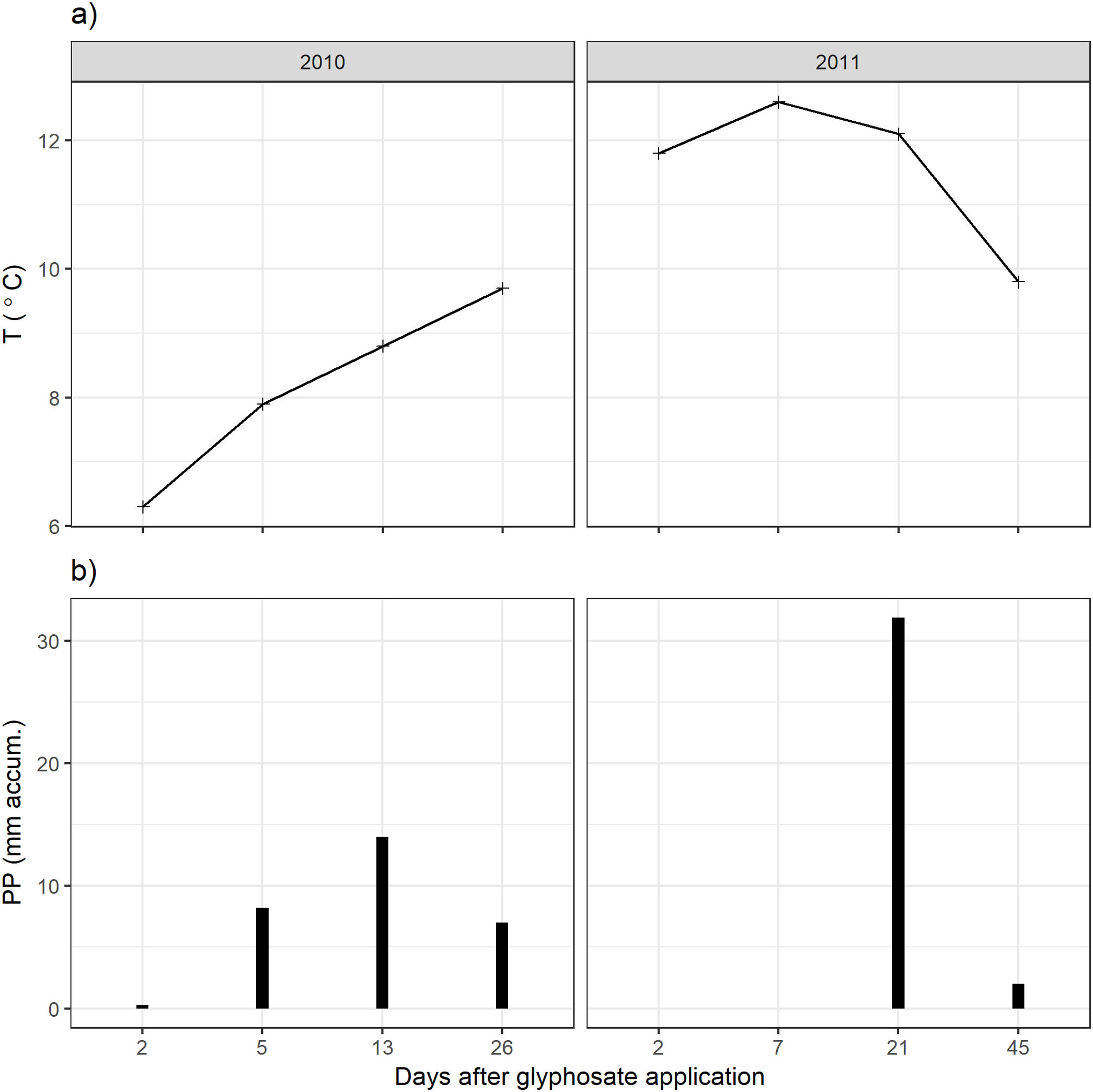
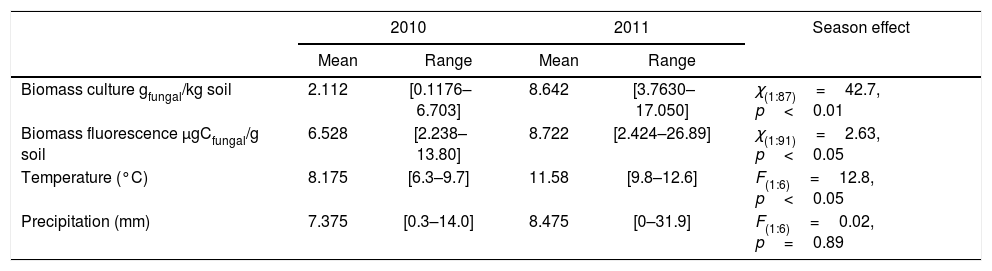
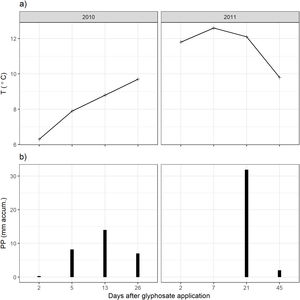
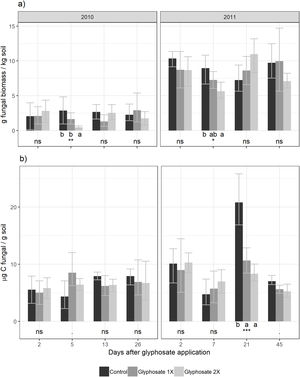
ResultsClimatic conditions and fungal biomass estimationThe climatic variables varied significantly between the seasons. The temperature was significantly higher during the second year of the study (Table 1, Fig. 1A). The rainfall showed no significant differences between the seasons but exhibited a wider variation during the second season reaching higher values (Table 1, Fig. 1B). Mean fungal biomass was 5.38g/kg (range=0.12–17.05g/kg) and 7.62μg C/g (range=2.24–26.89μg C/g), based on culture-dependent method and direct counting, respectively. It showed significant differences between the seasons considering both methods and was correlated with the climatic conditions (Table 1), resulting 409% and 133% higher during the second season of treatment with respect to the first season, with culture-dependent and direct estimation method, respectively.
Fungal biomass estimated with the culture-dependent method and epifluorescence microscopy, temperature and precipitation during 2010 and 2011.
| 2010 | 2011 | Season effect | |||
|---|---|---|---|---|---|
| Mean | Range | Mean | Range | ||
| Biomass culture gfungal/kg soil | 2.112 | [0.1176–6.703] | 8.642 | [3.7630–17.050] | χ(1:87)=42.7, p<0.01 |
| Biomass fluorescence μgCfungal/g soil | 6.528 | [2.238–13.80] | 8.722 | [2.424–26.89] | χ(1:91)=2.63, p<0.05 |
| Temperature (°C) | 8.175 | [6.3–9.7] | 11.58 | [9.8–12.6] | F(1:6)=12.8, p<0.05 |
| Precipitation (mm) | 7.375 | [0.3–14.0] | 8.475 | [0–31.9] | F(1:6)=0.02, p=0.89 |
For fungal biomass, the results of the season effect in the complete GLM model are shown.
For climatic variables, the results of one-way ANOVA comparing between seasons are shown.
For the culture-dependent method, the variability of fungal biomass within each year was not affected by any of the variables (treatment, days after herbicide application, temperature, precipitation, Likelihood Ratio Tests, p>0.13). For the direct estimation method, the variability of fungal biomass was not affected by any of these variables for the first year (Likelihood Ratio Tests, p>0.16). During the second year, it was significantly affected by the precipitation (Likelihood Ratio Test, p<0.001).
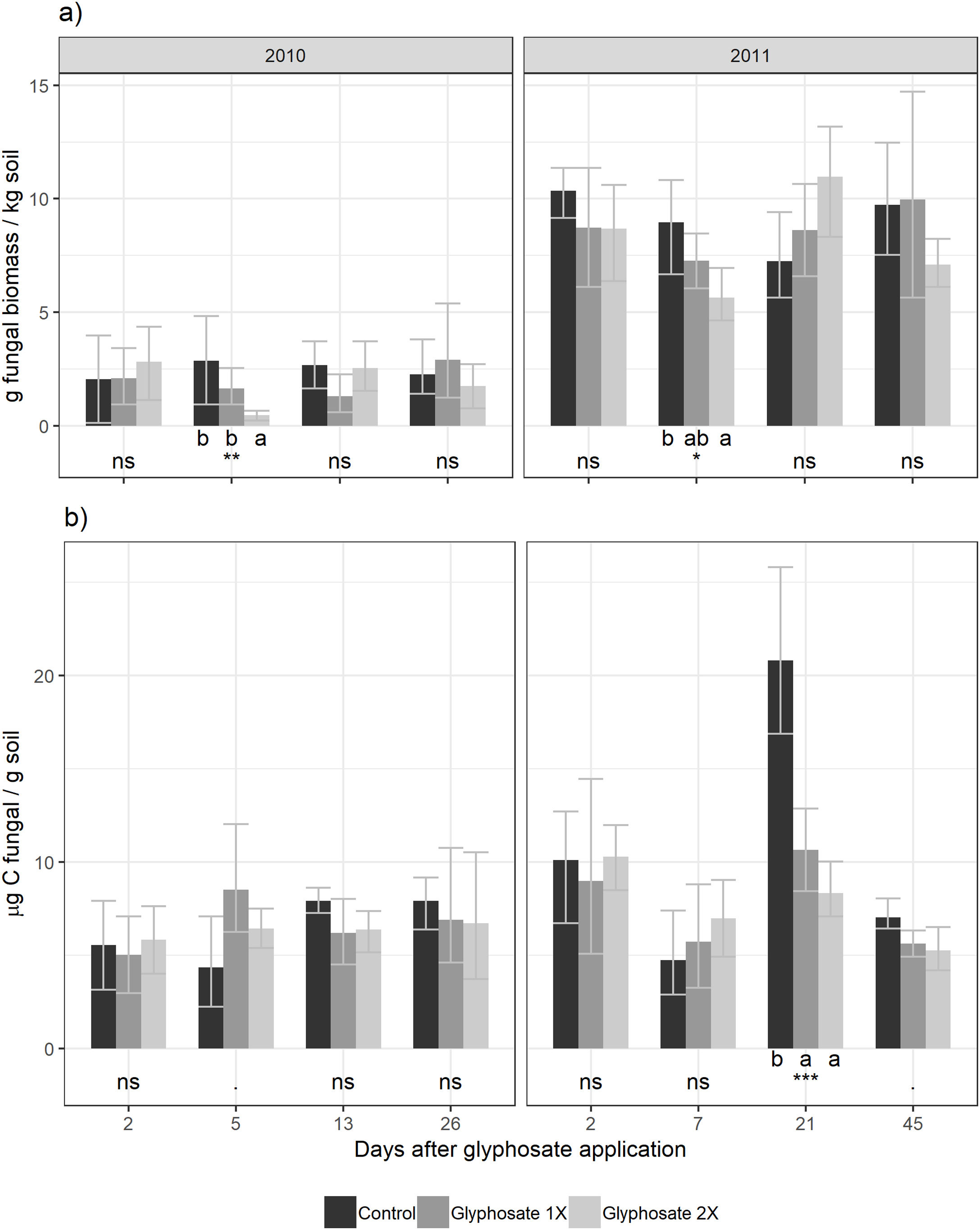
Five days after herbicide application, a decrease in the number of cultivable fungi was observed using the culture-dependent method (Tukey results shown in Fig. 2). This method detected temporal shifts in fungal biomass in plots treated with glyphosate during both seasons. Five days after glyphosate application, glyphosate 2× treatments showed a fungal biomass significantly lower than the control and Glyphosate 1× showed intermediate values. Fungal biomass showed no significant differences between the treatments for the rest of the dates.
Using the culture-independent method, an earlier and transitory stimulation effect of glyphosate on fungal biomass was observed 5 days after application during the first year (showing marginally significant differences, Fig. 2). During the second year, this trend was observed but was not statistically significant.
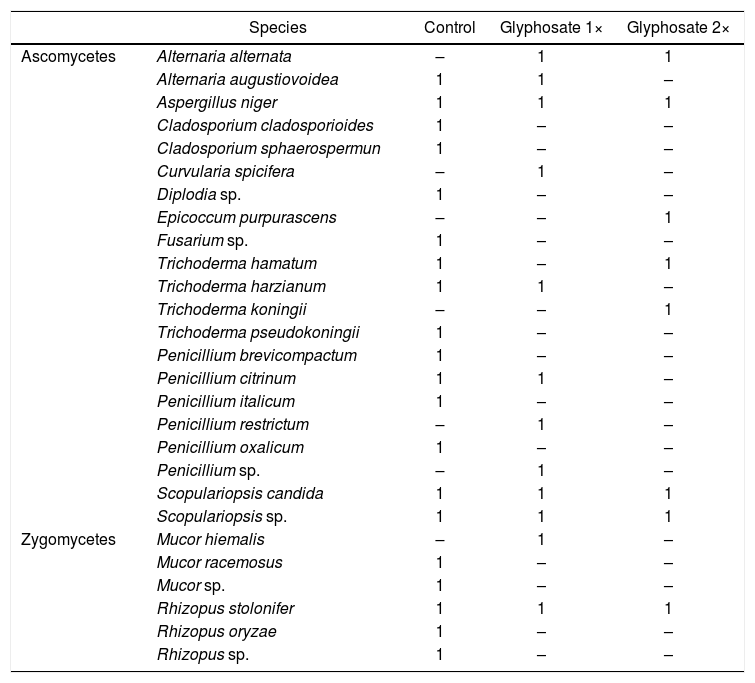
We found a significant correlation between both biomass estimation methods (t=2.58, d.f.=88, p=0.012) with a low coefficient of correlation (r=0.264, IC95%=[0.06–0.45]). Presence-absence data of the isolated fungal strains in each herbicide treatment are shown in Table 2. The number of strains isolated and identified was higher in the control plots.
Species and number of isolated strains from control, glyphosate 1× and glyphosate 2× plots.
| Species | Control | Glyphosate 1× | Glyphosate 2× | |
|---|---|---|---|---|
| Ascomycetes | Alternaria alternata | – | 1 | 1 |
| Alternaria augustiovoidea | 1 | 1 | – | |
| Aspergillus niger | 1 | 1 | 1 | |
| Cladosporium cladosporioides | 1 | – | – | |
| Cladosporium sphaerospermun | 1 | – | – | |
| Curvularia spicifera | – | 1 | – | |
| Diplodia sp. | 1 | – | – | |
| Epicoccum purpurascens | – | – | 1 | |
| Fusarium sp. | 1 | – | – | |
| Trichoderma hamatum | 1 | – | 1 | |
| Trichoderma harzianum | 1 | 1 | – | |
| Trichoderma koningii | – | – | 1 | |
| Trichoderma pseudokoningii | 1 | – | – | |
| Penicillium brevicompactum | 1 | – | – | |
| Penicillium citrinum | 1 | 1 | – | |
| Penicillium italicum | 1 | – | – | |
| Penicillium restrictum | – | 1 | – | |
| Penicillium oxalicum | 1 | – | – | |
| Penicillium sp. | – | 1 | – | |
| Scopulariopsis candida | 1 | 1 | 1 | |
| Scopulariopsis sp. | 1 | 1 | 1 | |
| Zygomycetes | Mucor hiemalis | – | 1 | – |
| Mucor racemosus | 1 | – | – | |
| Mucor sp. | 1 | – | – | |
| Rhizopus stolonifer | 1 | 1 | 1 | |
| Rhizopus oryzae | 1 | – | – | |
| Rhizopus sp. | 1 | – | – |
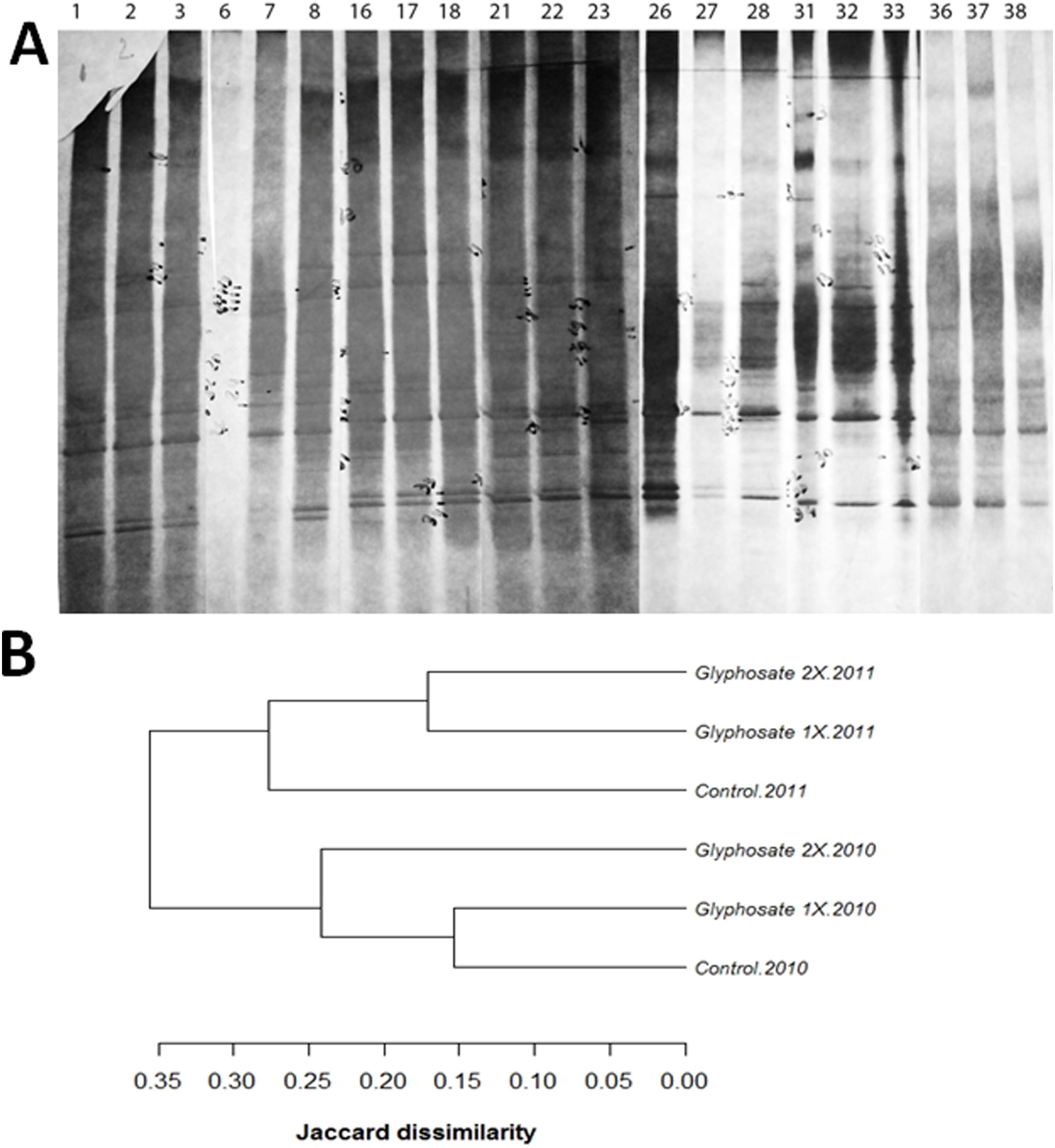
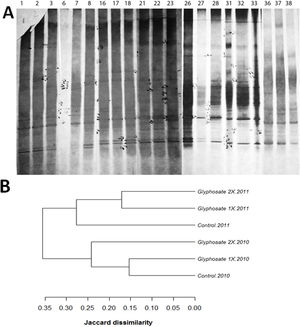
A total of 36 polymorphisms were revealed. Soil fungal profiles differed both in the presence/absence and the positions of the bands. Between 11 and 17 DGGE bands were detected in all samples with common positions in the upper part of the gel, and some minor bands in the middle part of the gel (Fig. 3). Matrix analysis reveals high number of polymorphisms in the treated plots and some unique bands characteristic of glyphosate treatments. Furthermore, no unique bands were found in the control soils (Fig. 3A). Of all the revealed polymorphisms, 16.66% were exclusively from glyphosate treated plots. Bands A9, A12 and A32 are unique for both Glyphosate 1× and 2× treatments and, bands A1 and A2 are unique for Glyphosate 2× treatment.
(A) Scanned image of silver stained DGGE gel (10% acrylamide, 30–60 denaturant) profiling the soil fungal communities exposed to glyphosate 1×, 2× and control, showing the position of individual bands for control (lanes1,6, 16, 21, 26, 31, 36), glyphosate 1× (lanes 2, 7, 17, 22, 27, 32, 37) and glyphosate 2× (lanes 3, 8, 18, 23, 28, 33, 38). (B) Dendrogram showing clustering analyses of the profiles, using the unweighted pairwise grouping method with mathematical averages. The analyses take into account the presence or absence of bands at certain positions in each lane of the gel.
The cluster analysis of the profiles revealed differences in the soil fungal communities after glyphosate exposure (Fig. 3B). The analysis showed a close correlation of DGGE profiles between the first and the second year of treatment. During the first year of study the control samples clustered in a group with glyphosate 1× samples from the same year. However, during the second year, the control was not in the same cluster as the two glyphosate treatments (Fig. 3B).
DiscussionThe effect of glyphosate on soil fungi has been extensively studied in vitro and in soil microcosm assays2,5,8,10,23,26,30,34,41,52. There are scarce and controversial reports related to the effect of this herbicide on soil fungi under field conditions, where multiple biological and physicochemical parameters are interacting4,22,48,50. An integrated approach, such as the one we present in this study, is intended to evaluate, simultaneously, ecological, structural and functional properties of soil fungi. The application of different methodologies enabled us to detect temporal shifts in fungal biomass in response to the application of glyphosate, changes in climatic conditions and the variation in the fungal community structure, obtaining contrasting results.
In a period of two years, no overall effect of glyphosate on the soil fungal biomass was observed, which was in accordance to reports that suggest that glyphosate has a low or null effect on fungal growth in the soil microcosm8,10. However, a more careful analysis of the data coming from the multiproxy approach employed enabled us to detect temporal shifts on fungal biomass after herbicide application.
The culture-dependent method detected a negative effect of glyphosate on soil fungal biomass after one week of its application in high doses, during both years of study. Growth might be different in Petri dishes compared to soil. The major limitation of this technique is thus, that it dramatically underestimates the microbial numbers and composition in the samples under study1. Soil is inhabited by a complex microbial consortium that contains a high proportion of low-abundance microorganisms43. It was interesting to observe that culturing methods do not capture most of the dominant microorganisms found in soils50. The number and composition of isolates recovered from control plots was higher than those from the treated soils (Table 2). Fungal community composition shows seasonal changes related to weather conditions36. Here we found changes in the community composition more closely related to glyphosate treatment than to the season. The negative effect on fungal biomass could be related to the effect of glyphosate exerted on low abundance fungal species.
On the other hand, the direct estimation method did not detect these falls in fungal biomass but showed sensitivity to precipitations. The direct method detected a peak in fungal biomass which can be associated to a peak of precipitation, highlighting the significance of climatic variables in the model. Changes in the soil fungal biomass were detected between years, and these changes would be more closely related to the environmental conditions46 than to glyphosate application29. The low rainfall in the first year of study could explain the difference in soil fungal biomass observed between years. Many other studies found that arid grasslands responded more strongly to wet years than to dry years24. Thus, in comparison to other ecosystems, fungal communities in grasslands show to be more resistant to short-term drought and respond rapidly after rainfall, increasing their biomass32.
Using a direct estimation method, an early and transitory stimulatory effect was observed in soil fungal biomass five to seven days after herbicide application. This glyphosate-induced stimulation effect was weak based on our statistical results; however, it is supported by the literature given it has been repeatedly observed in microbial communities under laboratory conditions17,22,34,41. Glyphosate has been shown to stimulate soil microbial activity, at least temporarily, suggesting that the herbicide is a source of nutrients22,60. Agrochemicals, including glyphosate, may represent an occasional source of C and nutrients for soil microorganisms, thus, they can be completely dissipated from the soil environment by either a single microbial species or the joint action of a microbial consortium60. Here, the temporal increase in fungal biomass could be related to glyphosate degradation and consumption, at least partially. Soil fungal metabolization of glyphosate has been confirmed both in vitro25,45 and in soil microcosms5. This behavior could be explained as an adaptive response frequently observed in microbial communities living in polluted and stressful environments39,47.
The number of isolated species from the control plots was higher than in the treated plots, suggesting a negative effect of glyphosate on the species richness of cultivable fungi. However, the molecular technique revealed that fungal species richness increased in most of the treated plots, and the pattern of the band analysis suggested a temporal shift in species composition. Each culture-dependent and culture-independent technique led to a unique microbial composition, sometimes completely different from those determined using molecular approaches. The microbial identities varied dramatically according to the analytical approach used1. Even more, a number of cultured microorganisms were not identified at all using molecular techniques1,50.
Molecular methods are useful analytical tools for evaluating the structure and function of microbial communities, as they jointly analyze the cultivable and non-cultivable members of the communities. The PCR-DGGE approach is frequently used in studies regarding soil microbial communities14,31,58. In this study, large numbers of polymorphisms were observed in the treated plots, suggesting that glyphosate has an effect on soil fungal diversity.
The results from the cluster analysis indicated that the effect of glyphosate is not immediate but can affect the structure of fungal communities in soils after repeated applications. Long term application of glyphosate may induce genetic changes on soil fungal species. Thus, the survivor communities are likely to have more mutations than normal and this would explain the large number of polymorphisms observed. Climatic conditions, agrochemical application and other agricultural practices induce shifts in the structure of soil fungal communities53. The stress generated by herbicide toxicity may alter the community structure of soil microorganisms60 through the activation of silenced genes and enhanced mutations18.
ConclusionsThe analysis of results suggested the convenience of studying the soil fungal community using combined approaches. The cultivation, direct estimation and molecular approaches complemented each other, allowing us to better understand the effect of glyphosate on soil fungal communities much better than any other method used alone. To our knowledge, this is the first study that combines all three approaches. In summary, the results of our studies are technique-dependent. Here we demonstrate a negative effect of glyphosate on soil fungal biomass at high doses, detected with the culture-dependent method. Furthermore, an early and transitory stimulatory effect on soil fungal biomass after glyphosate application was detected using the direct estimation method. We also found a negative effect of glyphosate on the species richness of cultivable fungi and changes in the molecular structure of soil fungal communities after double doses or long-term glyphosate application. In summary, our findings demonstrate an overall negative effect of glyphosate on soil fungal communities.
Conflict of interestThe authors declare that they have no conflicts of interest.
This study was funded by the Consejo Nacional de Investigaciones Cientificas y Técnicas (CONICET, Argentina) (Project number: PIP 11220130100280CO). The authors thank Rosemary Scoffield for language revision.