Cronobacter species are opportunistic pathogens associated with severe infections in neonates and immunocompromised infants. From January 2009 through September 2010, two cases of neonatal infections associated with Cronobacter malonaticus and one case associated with Cronobacter sakazakii, two of them fatal, were reported in the same hospital. These are the first clinical isolates of Cronobacter spp. in Argentina. The objective of this work was to characterize and subtype clinical isolates of Cronobacter spp. in neonate patients, as well as to establish the genetic relationship between these isolates and the foodborne isolates previously identified in the country. Pulsed-field gel electrophoresis analysis showed a genetic relationship between the C. malonaticus isolates from two patients. Different results were found when the pulsed-field gel electrophoresis patterns of clinical isolates were compared with those deposited in the National Database of Cronobacter spp.
Las especies del género Cronobacter son patógenos oportunistas asociados a infecciones graves en neonatos y niños inmunocomprometidos. Entre enero de 2009 y septiembre de 2010, en un mismo hospital se hallaron tres casos de infecciones neonatales, dos con desenlace fatal. Dos estuvieron asociados a Cronobacter malonaticus y uno a Cronobacter sakazakii. Estos son los primeros aislamientos clínicos de Cronobacter spp. notificados hasta el momento en Argentina. El objetivo de este trabajo fue caracterizar y subtipificar los aislamientos clínicos de Cronobacter spp. y establecer la relación genética entre dichos aislamientos y cepas de origen alimentario, previamente identificadas en el país. El análisis de los patrones de electroforesis en campo pulsado mostró una alta relación genética entre los aislamientos de C. malonaticus de dos de los pacientes. Los patrones de campo pulsado provenientes de las muestras clínicas resultaron diferentes a los patrones presentes en la Base de Datos Nacional de Cronobacter spp., de aislamientos de alimentos.
Cronobacter spp., formerly Enterobacter sakazakii, are members of the Enterobacteriaceae family. These orga-nisms were originally identified as yellow-pigmented Enterobacter cloacae. In 1980, they were classified as a new species, Enterobacter sakazakii. More recently, further phenotypic and molecular studies led to their reclassifica-tion into a new genus named Cronobacter, which comprises the type species Cronobaacter sakazakii and six other species, Cronobacter malonaticus, Cronobacter turicensis, Cronobacter muytjensii, Cronobacter dublinensis, Cronobacter condimenti and Cronobacter universalis7-9.
Cronobacter species are opportunistic pathogens that have been associated with severe infections such as septicemia, meningitis and necrotizing enterocolitis mainly in neonates and immunocompromised infants7,8. Severe neurological complications such as hydrocephalus, quadriplegia, brain abscesses and developmental delay have been observed in surviving patients.
Cronobacter spp. is widely spread in the environment and has been found in a variety of food, being the powdered infant formula (PIF) the main infection source described7. Furthermore, PIF contamination has been demonstrated, both as intrinsic to the product as well as during its preparation for consumption. Therefore, cleaning equipment, hospital environment and proceedings must be taken into consideration for surveillance and control of this pathogen5.
In Argentina, from 2005 through 2008, the mentioned pathogen was found in three different brands of imported PIF manufactured by companies from three different countries15. However, Cronobacter spp. infections have not been documented in our country so far.
In January 2009, a case of meningitis and septicemia in a preterm neonate twin was associated with Cronobacter spp. infection isolated from blood culture and cerebrospinal fluid (neonate A) at “Hospital Universitario Austral, Universidad Austral”, Buenos Aires, Argentina. In March 2009, a second case of septicemia, necrotizing enterocolitis with intestinal perforation in a preterm, neonate twin having low birth weight was registered in the same hospital (neonate B). This case presented co-infection by Cronobacter sp., Enterobacter cloacae, cogulase-negative staphylococci and Enterococcus faecalis (isolated from peritoneal liquid). Both patients had been fed with fluid and breast milk. An antibiotic treatment with vancomycin and amikacin was administered; however, both patients died. Fluid milk batches given to the neonates at the time of the infection were cultured in BACTEC bottles (Becton Dickinson, Franklin Lakes, NJ, USA) for 5 days but no pathogens were recovered.
To investigate possible environmental sources, swab samples were collected from the counter, tap, sink, floor, refrigerator, breast pump and trolley found in the feeders within neonatal intensive care units during the week following the patients’ diagnosis. These swabs, together with one breast milk sample (from neonate A's mother) were cultured in blood agar and chocolate agar (bioMérieux, Marcy l’Etoile, France). At the same time, these samples were cultured in brain heart infusion broth and thioglycolate broth (Britania, Buenos Aires, Argentina), and after enrichment, the cultures were transferred to solid agar plates for isolation of Cronobacter spp. This microorganism could not be recovered from any of the samples tested.
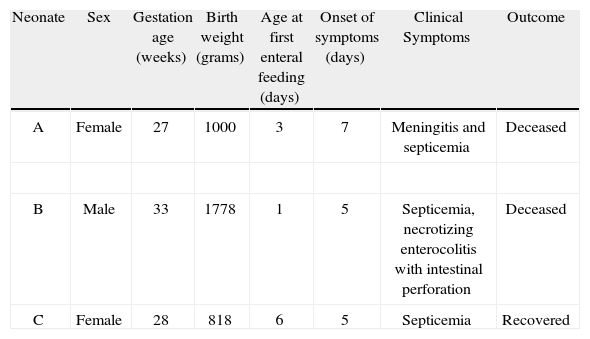
In September 2010, a third case of septicemia in a preterm neonate was associated with Cronobacter spp. infection, isolated from blood culture, at the same hospital (neonate C). The patient had been fed with breast milk through enteral feeding tubes. An antibiotic treatment with meropenem and amikacin was administered for fourteen days and the patient recovered. Clinical characteristics of the three cases described above are presented in Table 1.
Clinical characteristics of neonates A, B and C
| Neonate | Sex | Gestation age (weeks) | Birth weight (grams) | Age at first enteral feeding (days) | Onset of symptoms (days) | Clinical Symptoms | Outcome |
| A | Female | 27 | 1000 | 3 | 7 | Meningitis and septicemia | Deceased |
| B | Male | 33 | 1778 | 1 | 5 | Septicemia, necrotizing enterocolitis with intestinal perforation | Deceased |
| C | Female | 28 | 818 | 6 | 5 | Septicemia | Recovered |
The working protocol was approved by the Institutional Review Board at Universidad Austral (Comité de Evaluación Institucional de la Universidad Austral).
The objectives of this work were to characterize and subtype the clinical isolates of Cronobacter spp. from the neonate patients and to establish the genetic relationship between these isolates and the foodborne isolates that have been previously identified in the country.
Isolates were identified by conventional biochemical testing recommended for Ente robacte riaceae3,4,8 and by the use of commercial identifica tion biochemical API 20E galleries (bioMérieux) according to the manufacturers’ instructions. Utilization of malonate was determined to differentiate C. sakazakii from C. malonaticus8. Isolates were identified as C. sakazakii (neonate A), and C. malonaticus (neonates B and C).
Identification of the isolates as Cronobacter spp. was confirmed by PCR applying the method described by Mohan Nair and Venkitanarayanan for amplification of ompA sequences10. DNA was extracted by boiling a bacterial suspension in molecular quality water for 10 min, debris was centrifuged at 12,000 r.p.m. and the supernatant was used as template for the PCR assay15. PCR amplification of the ompA gene was positive for the three isolates tested.
The susceptibility of Cronobacter spp. isolates to antimicrobial agents was determined by the Kirby-Bauer disk diffusion test with the antibiotics recommended for Ente robacte riaceae2. The three isolates were susceptible to all the antimicrobial agents assayed (amoxicillin/clavulanic acid, ampicillin, cefotaxime, ceftriaxone, chloramphenicol, ciprofloxacin, gentamicin, nalidixic acid, tetracycline, trimethroprim-sulfamethoxazol, cefuroxime, cefixime, streptomycin, ceftazidime, cefoxitin, amikacin, piperacillin/tazobactam, imipenem), except for cephalothin. Our results were in agreement with previous studies7,11,15.
The genetic relationship among isolates was determined by pulsed-field gel electrophoresis (PFGE), considered a gold standard molecular epidemiology technique for subtyping bacterial isolates6. The PulseNet standardized protocol for Shigella sonnei for plug preparation and electrophoresis conditions was applied13. XbaI and SpeI were used as primary and secondary restriction enzymes, respectively. The PFGE patterns were analyzed using the BioNumerics software (version 4.0; Applied Maths, Sint-Martens-Latem, Belgium). The genetic profiles were compared and clustered applying the DICE coefficient and the Unweighted Pair Group Method with Arithmetic Averages, with a band position tolerance and optimization of 1.5%.
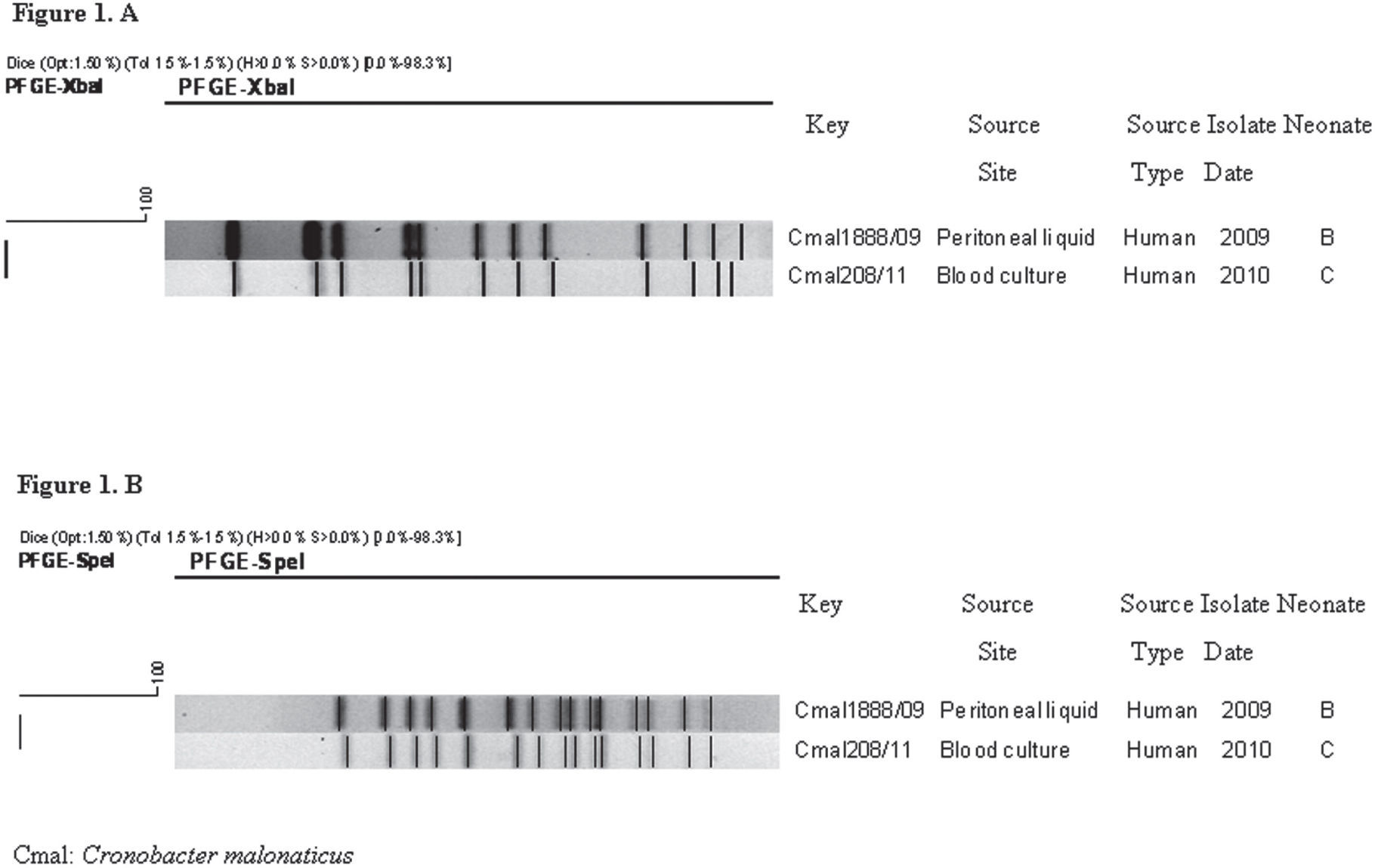
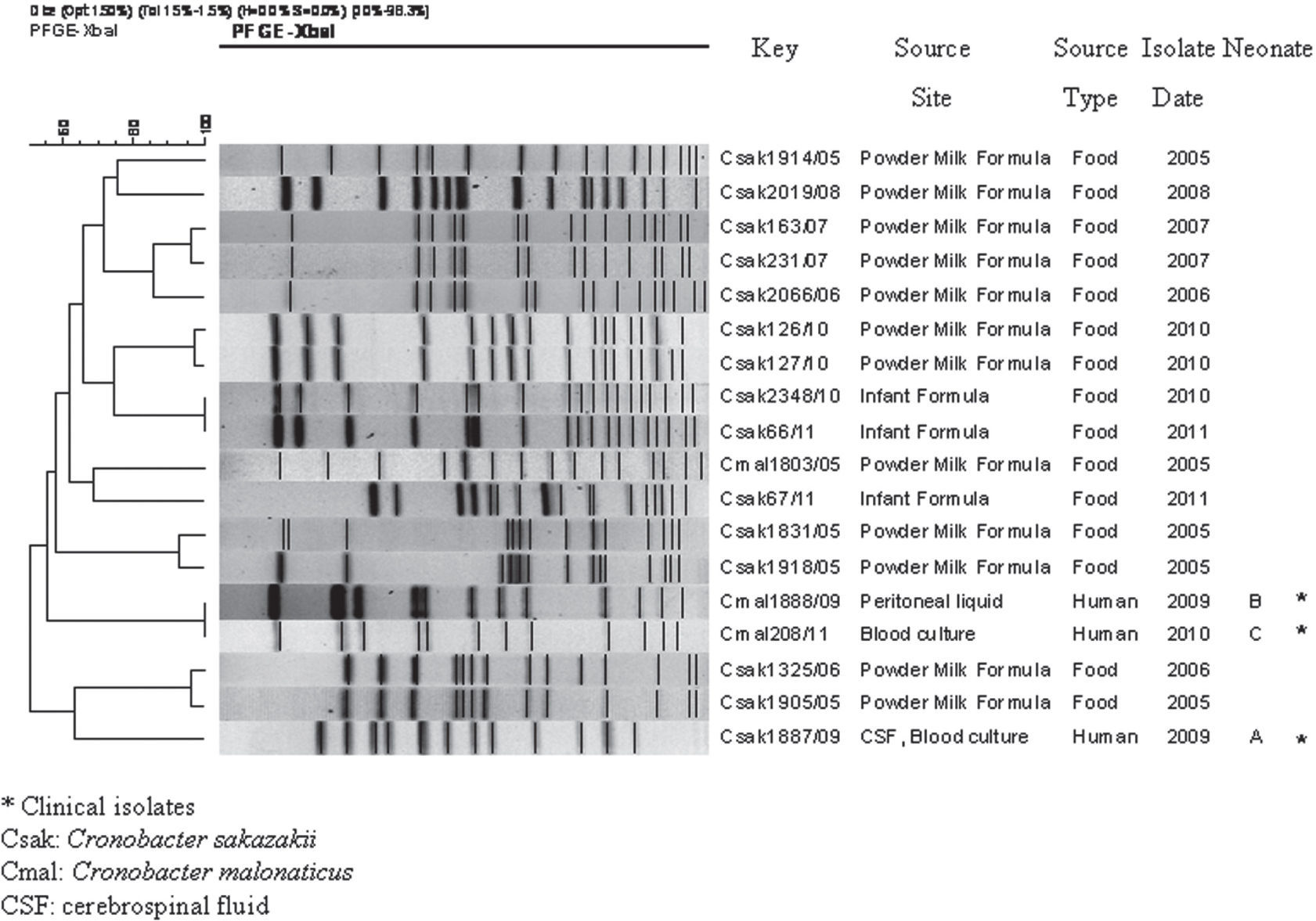
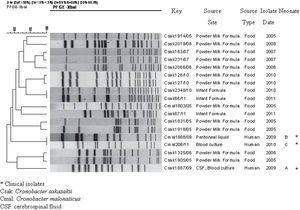
As expected, the PFGE pattern of the C. sakazakii strain isolated from neonate A, restricted with both enzymes, was different from the patterns of C. malonaticus isolates. XbaI PFGE analysis showed a high genetic relationship between the two isolates of C. malonaticus from neonates B and C (Fig. 1A). These results were confirmed using SpeI, with which both isolates showed the same PFGE pattern (Fig. 1B). XbaI PFGE patterns of the three isolates were found to be unique when compared with 15 PFGE patterns from 49 isolates (48 C. sakazakii and 1 C. malonaticus) deposited in the National Database of Cronobacter spp. (Fig. 2).
Clinical isolates of C. malonaticus obtained from neonates B and C were genetically related, suggesting that this strain could have persisted in an undetermined source or reservoir in the hospital from March 2009 to September 2010.
Although we only compared three isolates of C. malonaticus, there are previous studies1,11,14 which applied different PFGE protocols that demonstrated diversity among C. malonaticus isolates. This supports the idea that the genetic relationship between the two clinical isolates described herein was not due to limitations of PFGE subtyping or high clonality of the species analyzed.
Apart from the milk formula fed to the neonates, water outlets, medical equipment, surfaces and interpersonal contacts from the hospital environment could be the sources of the infection5,12. Despite the efforts for isolating the microorganism from the hospital environment, we were unable to identify the source of infection in the hospital setting. It is possible that the places selected and the procedures applied for sampling were not appropriate; furthermore, the culture techniques may not have been optimal for environmental and milk samples.
This is the first published report of Cronobacter spp. clinical isolates in Argentina. These findings stress the relevance of strengthening the surveillance of this pathogen in the susceptible population of neonates in Argentina, as a cause of potentially fatal infection.
As Cronobacter spp. has been isolated from different food sources, the study of this microorganism should also be enhanced in food and in industrial areas in order to prevent its spreading and also to improve food quality and safety. It is important to conduct adequate investigation in the hospital setting to detect possible environmental sources of Cronobacter spp. to prevent future infections.
Conflicts of interestThe authors declare that they have no conflicts of interest.
We thank Norma Binsztein and Raquel Terragno for providing expert advice and for encouraging the study of Cronobacter spp. in the country.