Leptospirosis is an endemic disease caused by Leptospira spp., a bacterium that affects animals and humans. In recent years, the number of reports of leptospirosis in wild animals has increased, which highlights the need to study the infectious agents in these animals. In this study, a duplex PCR for the detection of leptospiral DNA was performed on 50 kidney samples from bats, and a MAT (Microscopic Agglutination Test) for serological detection of anti-leptospiral antibodies was applied to 47 serum samples from bats from different regions of Buenos Aires Province, Argentina.
DNA was extracted using Chelex-100 and duplex PCR was performed by targeting the detection of genes secY and flaB, of pathogenic Leptospira spp. Of the 50 kidney samples, 3 were positive for Eumops sp. and Tadarida brasiliensis by duplex PCR. Of the 47 serum samples, 12 were positive for different serovars: Leptospira interrogans serovars Icterohaemorrhagiae, Cynopteri and Bataviae, and Leptospira borgpetersenii serovar Ballum. This is the first report of the detection of pathogenic leptospires by serology in bats belonging to the T. brasiliensis and Eptesicus furinalis species in Argentina. In addition, this is the first report of the detection of pathogenic leptospiral DNA by PCR in T. brasiliensis species. The detection of Leptospira spp. in these wild animals shows that they may play an important role as wildlife reservoirs of leptospires.
La leptospirosis es una enfermedad endémica causada por Leptospira spp., una bacteria que afecta a animales y a humanos. En los últimos años, el número de reportes de leptospirosis en animales silvestres ha aumentado, lo que resalta la necesidad de analizar los agentes infecciosos en estos animales. En este estudio, se aplicó una reacción en cadena de la polimerasa (PCR) dúplex para la identificación del ADN leptospiral en 50 muestras de riñones de murciélagos y la prueba de aglutinación microscópica (MAT) para la detección serológica de anticuerpos antileptospira en 47 muestras de suero de murciélagos de diferentes regiones de la provincia de Buenos Aires, Argentina. El ADN fue extraído usando Chelex-100 y la PCR dúplex estuvo dirigida a la detección de los genes secY y flaB de Leptospira spp. patógena. De las 50 muestras de riñón, tres resultaron positivas por PCR dúplex para Eumops sp. y Tadarida brasiliensis. De las 47 muestras de suero, 12 fueron positivas a diferentes serovares: Leptospira interrogans serovares Icterohaemorrhagiae, Cynopteri y Bataviae, y Leptospira borgpetersenii serovar Ballum. Este es el primer reporte de detección de leptospiras patógenas por serología en murciélagos pertenecientes a las especies T. brasiliensis y Eptesicus furinalis en Argentina. Además, también es el primero en la localización de ADN leptospiral por PCR en la especie T. brasiliensis. La identificación de Leptospira spp. en estos animales silvestres muestra que pueden desempeñar un papel importante como reservorios de leptospiras en la fauna silvestre.
Leptospirosis is a zoonotic disease caused by pathogenic strains of the genus Leptospira spp. Currently, the genus comprises 35 species34 with more than 300 serovars according to the composition of their lipopolysaccharide (LPS)21.
Recent reports show that leptospirosis has a global effect in terms of risk on human health27,36. In humans, the disease may vary from a flu-state to severe multiorgan involvement. In livestock animals, the disease can cause large economic losses due to reproductive failures, mainly in cattle production.
Transmission may occur by direct or indirect contact with urine or water contaminated with strains of pathogenic leptospires. Urine infected with leptospires is spread through the environment by reservoir hosts (mainly rodents), wild animals or domestic animals that can participate in the complex epidemiological cycle of leptospirosis as reservoirs and carrier hosts5,21,22.
Since the 1940s, most EID (Emerging Infectious Diseases) events have originated in wildlife, and of all EIDs zoonoses from wildlife represent the most significant ones, as they imply a growing threat to global health16. Zoonotic pathogens have been identified in carnivores, rodents, primates, bats and other wild animals39 and their incidence has continued to increase16.
Due to their abundance and broad spatial distribution, bats have been involved in numerous EID events, and are increasingly recognized as important reservoir hosts for bacteria, rickettsiae and viruses (alphaviruses, flaviviruses, rhabdoviruses and arenaviruses)8,23,28. However, the role of bats as hosts of bacterial zoonotic agents is almost unknown.
In many regions of Latin America, people live in precarious neighborhoods establishing ideal ecological conditions for the transmission of zoonotic diseases, mainly from synanthropic animals such as rodents and bats. In this context, bats have been postulated as possible sources of leptospiral infection11,40.
Leptospira interrogans, a typical rodent-borne Leptospira species has been detected in frugivorous and insectivorous bats, and therefore, the transmission between rodents and bats has been suggested19,25. Thus, indirect transmission of Leptospira spp. to humans may also occur through bat-borne pathogenic strains and other animal-associated leptospires, in particular rodents that reside or forage under bat roosts25,33.
However, direct transmission has not been evidenced38. Currently, Argentina has few data of leptospirosis in bats, although there is existing evidence of the important role that these animals play in the transmission of different zoonotic diseases.
The aim of this work was to detect leptospiral DNA by PCR and anti-Leptospira antibodies by serology in bats from Argentina.
Materials and methodsSampling areaBats, dead and stored at 4–5°C, were received at the Urban Zoonoses Laboratory of the Ministry of Health of Avellaneda (Buenos Aires Province, Argentina) and Pasteur Zoonosis Institute (Buenos Aires City, Argentina).
A total of 50 animals (collected between 2015 and 2018) were used in this study: 44 samples belonged to the family Molossidae and 6 samples belonged to the family Vespertilionidae (Table 1). In each case, both kidneys were extracted with sterile dissection forceps and scissors, placed in a dry sterile Eppendorf tube and preserved at −20°C. These samples were collected from different locations of Buenos Aires Province (Fig. 1, supplementary material), and were received in the Laboratory of Leptospirosis, OIE Reference Center, located at the Institute of Pathobiology (National Institute of Agricultural Technology, Castelar-INTA Buenos Aires, Argentina) for their processing. In addition, 47 serum samples (collected between 2012 and 2013) were submitted to detect anti-Leptospira antibodies.
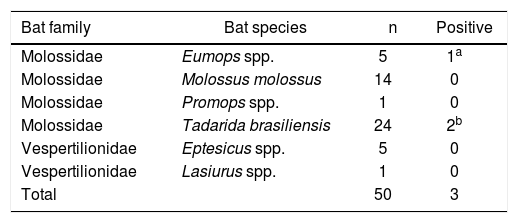
Bat species analyzed in the Laboratory of Leptospirosis (INTA Castelar) and positive cases for pathogenic Leptospira spp.
| Bat family | Bat species | n | Positive |
|---|---|---|---|
| Molossidae | Eumops spp. | 5 | 1a |
| Molossidae | Molossus molossus | 14 | 0 |
| Molossidae | Promops spp. | 1 | 0 |
| Molossidae | Tadarida brasiliensis | 24 | 2b |
| Vespertilionidae | Eptesicus spp. | 5 | 0 |
| Vespertilionidae | Lasiurus spp. | 1 | 0 |
| Total | 50 | 3 |
n: number of samples.
DNA was extracted from the kidney samples using 150μl of 5% solution of Chelex-100®15 (Biorad USA). Incubation was performed at 56°C for 20min and 100°C for 8min. Later, the samples were centrifuged at 10000rpm for 5s and 100μl of the supernatant was collected in 1.5ml tubes and stored at −20°C until use.
Duplex PCR was performed using primers G1:G2 (G1: 5′-CTGAATCGCTGTATAAAAGT-3′; 5′-G2:GCAAAACAAATGGTCGGAAG-3′), and B64I-B64II (B64I: 5′-CTGAATTCTCATCTCAACTC-3′ and B64II: 5′-GCAGAAATCAGATGGACGAT-3′) (modified from Gravekamp et al., 1993)13 which amplify a fragment of 285bp and 563bp from secY and flaB respectively13. The PCR reaction was carried out according to Grune et al. (2015)15. Amplification products were analyzed by electrophoresis in 2% agarose gel stained with ethidium bromide, followed by exposure to UV light (Uvi Tec transilluminator BTS-20.M). Amplicon sizes were estimated using a 100bp ladder (Embiotec, Buenos Aires, Argentina).
Microscopic Agglutination Test (MAT)The serum samples from 47 bats were tested against a panel of 23 serovars of Leptospira spp. by MAT. The panel was composed by the following serovar-reference strains: Australis-Ballico; Autumnalis-Akiyami A; Castellonis-Castellon III; Bataviae-Swart; Canicola-Hond Utrecht IV; Celledoni-Celledoni; Cynopteri-3522C; Djasiman-Djasiman; Grippotyphosa-Mosvka V; Hebdomadis-Hebdomadis; Icterohaemorrhagiae-RGA; Javanica-Veldrat Batavia 46; Louisiana-LSU 1945; Manhao-Manhao; Mini-Sari; Panam-CZ 214K; Pomona-Pomona; Pyrogenes-Salinem; Ranarum-ICF; Sarmin-Sarmin; Wolffi-Wolffi 3705; Shermani-LT 821 and Tarassovi-Perepelicin.
Initial serum dilution in phosphate buffered saline (PBS buffer) was 1:25.
ResultsDuplex PCR was performed on 50 samples and a positive result was observed in 3 of them. We observed the amplification of a 285bp fragment without amplification of the negative control. DNA integrity of negative samples was controlled by electrophoresis in 1% agarose gel and PCR for the 16S rRNA gene (data not shown).
The morphological analysis of the bats coincides with the taxonomic codes published in Barquez et al., and showed that the positive specimens belonged to the Eumops (1 animal) and Tadarida brasiliensis (2 animals), both from family Molossidae (Table 1).
Of the 47 serum samples tested by MAT, only 12 were positive for L. interrogans serovar-reference strains: Icterohaemorrhagiae-RGA, Bataviae-Swart, Castellonis-Castellon III, Cynopteri-3522 with titers greater than or equal to 1:25 (Table 2).
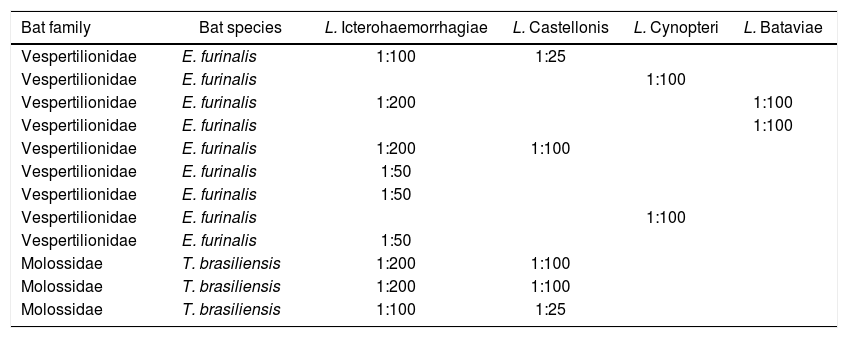
MAT titers to detect pathogenic Leptospira serovars in bats.
| Bat family | Bat species | L. Icterohaemorrhagiae | L. Castellonis | L. Cynopteri | L. Bataviae |
|---|---|---|---|---|---|
| Vespertilionidae | E. furinalis | 1:100 | 1:25 | ||
| Vespertilionidae | E. furinalis | 1:100 | |||
| Vespertilionidae | E. furinalis | 1:200 | 1:100 | ||
| Vespertilionidae | E. furinalis | 1:100 | |||
| Vespertilionidae | E. furinalis | 1:200 | 1:100 | ||
| Vespertilionidae | E. furinalis | 1:50 | |||
| Vespertilionidae | E. furinalis | 1:50 | |||
| Vespertilionidae | E. furinalis | 1:100 | |||
| Vespertilionidae | E. furinalis | 1:50 | |||
| Molossidae | T. brasiliensis | 1:200 | 1:100 | ||
| Molossidae | T. brasiliensis | 1:200 | 1:100 | ||
| Molossidae | T. brasiliensis | 1:100 | 1:25 |
The morphological analysis of the bats showed that the 9 positive specimens belonged to species Eptesicus furinalis (family Vespertilionidae), and 3 to T. brasiliensis (family Molossidae).
DiscussionDifferent research works have estimated that 60% of the emerging pathogens that affect humans are zoonotic, and that 70% of them originate in wildlife16. This demonstrates the importance of wild animals in several epidemiological cycles of pathogens included in human and domestic animal diseases.
In the last years, the expansion of urban areas, the increase in population density, the worldwide trips and traffic of wild fauna are some of the causes for the rise in contact between wild animals and humans36. Many wild animals carry leptospires, which they can spread into the environment by their elimination in the urine37.
In Argentina, pathogenic strains of Leptospira spp. were isolated and identified by molecular genotyping (MLVA) from wild boars (Sus scrofa)6, rodents (Rattus norvegicus, Rattus rattus)14, arboreal squirrels (Callosciurus erythraeus)12, gray foxes (Lycalopex griseus)32, skunks (Conepatus chinga)31, weasels (Didelphys albiventris)5, and armadillos (Chaetophractus villosus)1; strains of serovar Icterohaemorrhagiae have been repeatedly isolated from these wild animals1,2.
Within the wild animals, bats (Chiroptera) have been implicated in numerous emerging infectious disease events and are increasingly being recognized as important reservoir hosts for rickettsiae, bacteria and viruses8,23,28.
In several Latin American countries, the presence of pathogenic leptospiral DNA in bats has been detected by PCR. Mexico reported leptospiral DNA of Leptospira noguchii and Leptospira weilii in bats11, being the first country to detect species of pathogenic leptospires in flying mammals in North America. Peru reported L. interrogans, Leptospira borgpetersenii, Leptospira kirschneri and the intermediate species Leptospirafainei7,25 (view Fig. 2, supplementary material).
Due to changes in their habitat and expansion of urban areas, the distribution of these animals has changed and has increased contact between humans and these animals. However, little is known about the role of bats as hosts for bacterial zoonotic diseases8.
With regard to the serological techniques, the MAT is widely used to detect antibodies against leptospires, as it is considered the gold standard test for the OIE (World Organization for Animal Health). Positive serology has been reported in Argentina from wild animals such as the large hairy armadillo (C. villosus)17, the water buffalo (Bubalus bubalis)18 and the nutria (Myocastor coypus)24.
The presence of anti-Leptospira antibodies in bats in some countries of Latin America such as Trinidad and Tobago, Grenada and Brazil, has been detected with titers varying from 1:100 to 1:1600 against different serovars of Leptospira spp9,26,35,40 (Fig. 2, supplementary material). As in many other diseases, the presence of seroreactivity does neither imply clinical disease in these animals nor a role as maintenance hosts of this pathogen, but rather evidences that these bats have been in contact with leptospires in their environment.
Currently, there is scant information in Argentina of Leptospira spp. in bats. The only available report is from Ramirez29, who reported the presence of leptospiral DNA in kidney samples from bats of suburban areas (northern region of Argentina), in individuals belonging to Molossus rufus (Black mastiff bat) and Eumops patagonicus (Patagonian dwarf bonneted bat), both from family Molossidae, and Myotis albescens (Silver-tipped myotis) family Verspertilionidae.
In this work, we report the first detection of leptospiral DNA of pathogenic leptospira in T. brasiliensis, a bat species from Argentina belonging to family Molossidae.
Furthermore, we also report the first serological detection in Argentina in T. brasiliensis and E. furinalis (family Vespertilionidae). We found antibodies against leptospiral serovars Icterohaemorrhagiae, Ballum, Cynopteri and Bataviae (Table 2). This finding constitutes a milestone in the leptospiral research in our country as there have been no serological data in bats to the present date. Furthermore, this is the first report in E. furinalis species in South America, and the first report in bats of urban areas from the central region of our country.
It is interesting to highlight that these serovars are usually associated with other animal hosts, such as rodents (Icterohaemorrhagiae)1, canines (Icterohaemorrhagiae and Castellon)1, weasels (Didelphys albiventris) (Bataviae)1 and pigs (Castellon, Icterohaemorrhagieae)2. In many regions, cattle are infected by Leptospira borgpetersenii serovar Hardjo bovis (serovar usually associated with bovines) and serovars Pomona and Grippotyphosa2. The high diversity of serovars found in domestic and wild animals such as bats, reveals the high-genetic diversity of this genus. This is congruent with Lei et al.20, who proposed that leptospires found in bats are structured by geography rather than by the phylogenetic relationships of their hosts.
T. brasiliensis and E. furinalis are widely distributed in South America. While Tadarida spp. is known to be distributed from western Venezuela, Colombia, Ecuador, Peru, Bolivia, Paraguay, southern Brazil, Chile and Argentina30, Eptesicus spp. has a wide distribution in the neotropical region except the western slope of the Andes Mountains in Peru, Bolivia and Chile3,10. In Argentina T. braziliensis has been sighted throughout the country, except in Chaco, Corrientes and Tierra del Fuego Provinces. In addition, E. furinalis is distributed from the north of Argentina to La Pampa Province4.
The Urban Zoonosis Laboratory in the province of Buenos Aires received 513 animals from family Molossidae and 76 from family Vespertilionidae between 2015 and 2018, which shows the need to continue studying these families of bats in Argentina. These finding of leptospires in the species T. brasiliensis and E. furinalis is of great importance, since they are two of the most frequently seen species and of which there are numerous records within these bats circulating in the country3.
We also highlight that the isolation and genomic studies of the leptospires circulating in the sampling areas are necessary tools for the identification and confirmation that these pathogenic strains could be circulating among different wildlife species knowing that the distribution of these wild animals is important from an ecological approach, but even more so from an epidemiological point of view since it allows us to understand the distribution of many zoonotic agents, including the agent Leptospira spp.
Conflict of interestThe authors declare that they have no conflicts of interest.
We thank M. Ayelen Lutz, PhD, in Natural Sciences, Argentine Bat Conservation Program (ABCP) for her valuable input as well as for her logistical support. We are also grateful for the sponsorship of the National Institute of Agricultural Technology (INTA), National Animal Health Program PNSA 1115052. Vanina Saraullo has a CONICET Doctoral Scholarship.