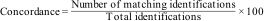Salmonella spp. is one of the most important zoonotic pathogens that causes foodborne diseases. It is divided into two species (Salmonella bongori and Salmonella enterica) including around 2600 serovars, being S. enterica serovar Enteritidis one of the most frequent in Argentina. Serovar identification is generally conducted by somatic and flagellar agglutination sera assays, and molecular biology techniques can also be performed. As efforts are being made worldwide to differentiate Salmonella serovars, our aim was to evaluate the utility of two specific biomarkers, previously reported for differentiating S. Enteritidis with MALDI-TOF MS. A panel of 105 S. enterica local isolates, belonging to different serovars and characterized by sera agglutination and PCR, was included in this study. Two specific S. Enteritidis biomarkers, at m/z 3016±3Da and 6034±3Da, were visually detected showing a sensitivity of 54% and 98%, respectively, and a specificity of 100% for both peaks. Concordance between serotyping and identification by PCR of S. Enteritidis and the blind search of biomarkers in a subset of isolates was 98%. Visual detection of these S. Enteritidis biomarkers using MALDI-TOF MS can be used as a fast and easy screening method for serovars differentiation at the microbiology clinical laboratory.
Salmonella spp. representa uno de los patógenos zoonóticos causantes de enfermedades transmitidas por alimentos más importantes a nivel mundial. Este género se divide en dos especies (S. bongori y S. enterica) que abarcan en conjunto aproximadamente 2.600 serovariedades. Salmonella enterica serovar Enteritidis es una de las serovariedades más frecuentes en Argentina. Su serotipificación se realiza habitualmente mediante aglutinación con sueros, y asimismo se pueden utilizar técnicas de biología molecular. Dada la importancia de diferenciar serovariedades de Salmonella, nuestro objetivo fue evaluar la utilidad de dos biomarcadores específicos previamente reportados para diferenciar S. Enteritidis con MALDI-TOF MS. Se incluyó un panel de 105 aislamientos de Salmonella enterica locales, de diferentes serovariedades y caracterizados por aglutinación con sueros, por PCR o por ambos métodos. En esta colección se detectaron visualmente dos biomarcadores específicos de S. Enteritidis, de 3.016±3Da y 6.034±3Da, con una sensibilidad del 54% y del 98%, respectivamente, y una especificidad del 100%. La concordancia entre la serotipificación/identificación por PCR de S. Enteritidis y la búsqueda a ciegas de los biomarcadores fue del 98%. La detección visual de estos biomarcadores específicos de S. Enteritidis con MALDI-TOF MS puede usarse como método de cribado para su diferenciación de otras serovariedades en el laboratorio de microbiología clínica.
Salmonella, a genus belonging to Enterobacteriaceae family, is a gram negative, facultative aerobic bacillus, divided into two species (Salmonella bongori and Salmonella enterica, the latter divided in six subspecies) and includes around 2600 serovars. It is known as one of the most important zoonotic pathogens that causes foodborne diseases producing diarrhea, vomiting and bacteremia, among others8,17.
Of 725 strains identified in 2023 in our Reference National Laboratory for Salmonella within the framework of National Diarrhea Network, the most frequent serovars were S. Typhimurium (21.65%), S. Enteritidis (16.82%), S. Paratyphi B (12.41%), S. Newport (6.06%) and S. Oranienburg (4.41%). Serovar identification is generally conducted by somatic and flagellar agglutination sera assays1,5 as the gold standard methodology, with a turn-around-time (TAT) of approximately 7 days. Furthermore, molecular biology techniques such as conventional polymerase chain reaction (PCR)4,6,7, real-time PCR9 or sequencing16, with shorter TATs, can be performed.
MALDI-TOF MS is currently used for the identification of microorganisms, only allowing characterization at the genus level for Salmonella15. Some researchers described different serovar-specific biomarkers for S. Enteritidis3,18, S. Typhimurium18 and S. Typhi11 among others. Yang et al.18, found two S. Enteritidis specific biomarkers (mass peaks at 3018±1Da and 6037±1) analyzing 132 isolates after protein extraction with organic solvents. As efforts are being made worldwide to differentiate Salmonella serovars11,14, our aim was to evaluate the utility of these two specific S. Enteritidis biomarkers with MALDI-TOF MS18, using a local strain collection of different Salmonella serovars isolated in Argentina.
Materials and methodsIsolates for retrospective studyA panel of 105 isolates deposited in our strainer was included in this study for biomarker detection from colonies (direct method) (Table S1). Strains were recovered from different sources: 93 human clinical specimens (67 stool samples, 17 blood samples, 4 urine samples, 2 rectal swabs, 1 bile fluid, and 2 samples of unknown origin), 3 animal samples (meet/chicken) and 2 environmental samples; the origin for 7 samples was unknown (data not collected). Our set contained 50 S. Enteritidis and 55 non-S. Enteritidis isolates (16 S. Typhimurium, 8 S. Paratyphi B, 5 S. Anatum, 6 S. Newport, 4 S. Stanley, 2 S. Agona, 3 S. Typhi, 3 S. Saintpaul, 3 S. Sandiego, 3 S. Oranienburg and 2 S. Infantis). All isolates belong to the National Diarrhea Network for the surveillance of enteropathogens (Red Nacional de Diarreas y Patógenos Bacterianos de Transmisión Alimentaria, Argentina) and were recovered on Salmonella-Shigella Agar (SS) (Britania, Argentina). In addition, 6 S. Enteritidis and 7 non-S. Enteritidis isolates were analyzed for biomarkers visual detection after protein extraction with organic solvents (extraction method).
Phenotypic and genotypic characterization of isolatesAfter isolate recovery on SS Agar, strains were grown on Tryptein Soy Agar (TSA) (Britania, Argentina) and tested by Triple Sugar Iron Agar (Britania, Argentina) and Lisine Decarboxilase (Britania, Argentina) assays. Salmonella typical colonies were tested by antisera agglutination, according to the White-Kauffmann-LeMinor scheme, for serotyping13, and identified by MALDI-TOF MS. PCR assays were performed for somatic (O:8, O:4, O:9, O:7)7 and flagellar antigens (H:d, H:b, H:e,h, H:i, H:G, H:1,2 and Sdf I region)6,7 characterization, as implemented for routine serovar discrimination at our laboratory. Some isolates were also characterized by whole genome sequencing (WGS) at Unidad Operativa Centro Nacional de Genómica y Bioinformática, as shown in Table S1.
MALDI-TOF MS biomarker detection protocol from colonies grown on solid culture media (direct method)Pure cultures were obtained on TSA plates after overnight incubation at 37°C, and fresh colonies were spiked on the MALDI steel plate (MSP 96 target ground Steel; Bruker Daltonics, Germany) by triplicate and covered with 1μl of α-cyano-4-hydroxycinnamic acid (HCCA) matrix (Bruker Daltonics, Germany)2. Some S. Enteritidis isolates (n=8) were tested directly from SS Agar for biomarker visual detection. After air drying, the MALDI plate was introduced into the MicroFlex LT spectrometer version 3.4 (Bruker Daltonics, Germany) for spectra acquisition.
MALDI-TOF MS protein extraction protocol (extraction method)Protein extraction was performed as previously described2. Briefly, a loopful of Salmonella cells recovered in TSA was suspended in 300μL of deionized water, and inactivated with 900μL of ethanol (Sigma Aldrich, Germany). The suspension was centrifuged at 13600×g for 2min and the dried pellet was resuspended in 50μL of 70% formic acid (Sigma Aldrich, Germany) and 50μL of acetonitrile (Sigma Aldrich, Germany). After centrifugation at 13600×g for 2min, 1μL of supernatant was spotted onto the MALDI steel plate and overlaid with 1μL of HCCA after air drying. The MALDI plate was then introduced into the MicroFlex LT spectrometer version 3.4 (Bruker Daltonics, Germany) for spectra acquisition. Spectra were analyzed using commercially available Biotyper 3.1 version 12 database.
Blind biomarkers search in Salmonella spp. isolates from coloniesWe blindly searched for the selected biomarkers in 55 samples received at our service between December 2023 and January 2024, identified at genus level as Salmonella spp. We decided to apply the direct method, as (despite intensities differences) spectra general quality from colonies and the extraction method in the retrospective study was similarly acceptable. In addition, the direct method is simpler and requires fewer reagents than the protein extraction. We processed isolates from TSA by the direct method, as described in “MALDI-TOF MS biomarker detection protocol from colonies grown on solid culture media (direct method)” section. We searched for biomarker presence/absence after acquisition of spectra, and later, we compared the results with serovars obtained by serology or PCR assays, to determine the concordance between methods, according to the following formula:
We considered a matching identification when the serovar result obtained by serology/PCR was the same as the one obtained by biomarker visual detection.
Spectra acquisitionSpectra were acquired automatically using FlexControl 3.4 software (Bruker Daltonics, Germany). Spectrometer parameters were set as follows (default settings): mass range: 1960–20000Da, detection gain: 2702V, frequency: 60Hz, ion source 1: 19.97kV, ion source 2: 17.73kV.
Statistical analysisAfter baseline subtraction and smoothing of the raw spectra with FlexAnalysis 3.4 software (Bruker Daltonics, Germany), we performed visual analysis searching for biomarkers of S. Enteritidis at m/z ∼3018Da and ∼6037Da previously reported by Yang et al.18. Peaks presence/absence was visually investigated in every replicate. When the biomarker was present in 2/3 or 3/3 replicates, the isolate was identified as S. Enteritidis, and as non-S. Enteritidis when the biomarker was present in 1/3 replicates or not present. Each signal was evaluated individually.
Moreover, the analysis with ClinPro Tools vs. 3.0 software (Bruker Daltonics, Germany) was performed with spectra analyzed for blind biomarkers search10. Spectra were loaded into the software and characteristic peaks among different classes (Class 1: S. Enteritidis; Class 2: non-S. Enteritidis) were selected by the available statistical tests. After using the “Peak Statistic tool”, p-values for the Anderson–Darling (AD) and the Wilcoxon or Kruskal–Wallis (W/K–W) test were evaluated for the best ten discriminating peaks, and the area under the receiving operating curve (AUCROC) was analyzed for each peak.
ResultsPhenotypic and genotypic characterizationAll isolates were identified as Salmonella at the genus level using MALDI-TOF MS. Twenty-three isolates were characterized solely by antisera agglutination, 17 serovars were determined by PCR assays, and 65 isolates were analyzed by both methods simultaneously (Table S1). Additionally, three isolates (1 S. Enteritidis, 1 S. Typhi and 1 S. Stanley) were analyzed by WGS.
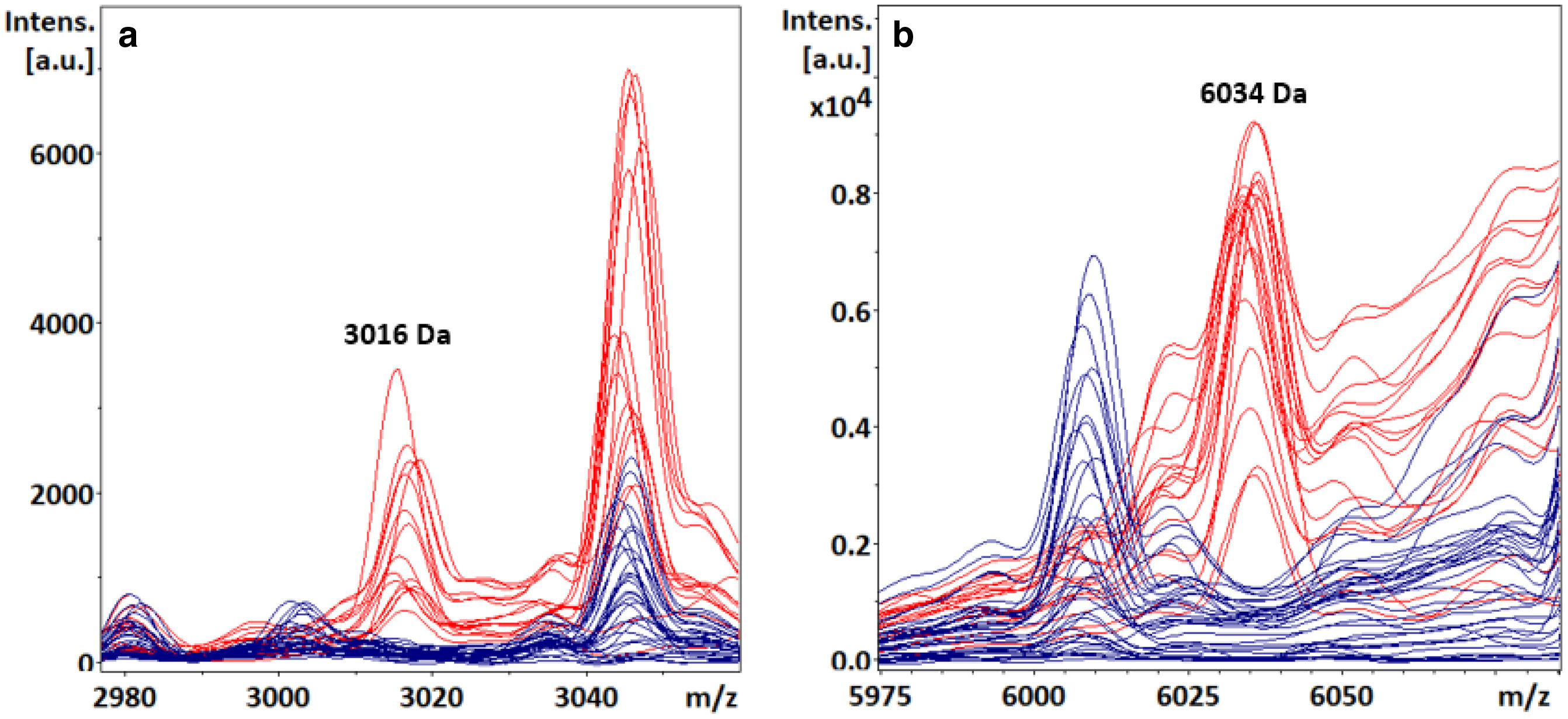
Visual detection of S. Enteritidis specific biomarkers with FlexAnalysis (direct and extraction method)In the retrospective study, both S. Enteritidis specific biomarkers were visually detected from colony (direct method) at m/z 3016±3Da (Fig. 1a) and 6034±3Da (Fig. 1b), showing a sensitivity of 54% and 98%, respectively, and a specificity of 100% for both peaks (Table 1) from TSA. We did not detect these peaks when processing isolates from SS Agar. Signal intensities in S. Enteritidis isolates ranged from 207 arbitrary units (a.u.) to 4983 a.u. for the ∼3016Da peak, and from 120 a.u. to 11 750 a.u. for the ∼6034Da peak. These two biomarkers were not detected in non-S. Enteritidis isolates, although some overlapping signal (noise) was observed at the m/z values evaluated, that did not represent a real mass peak.
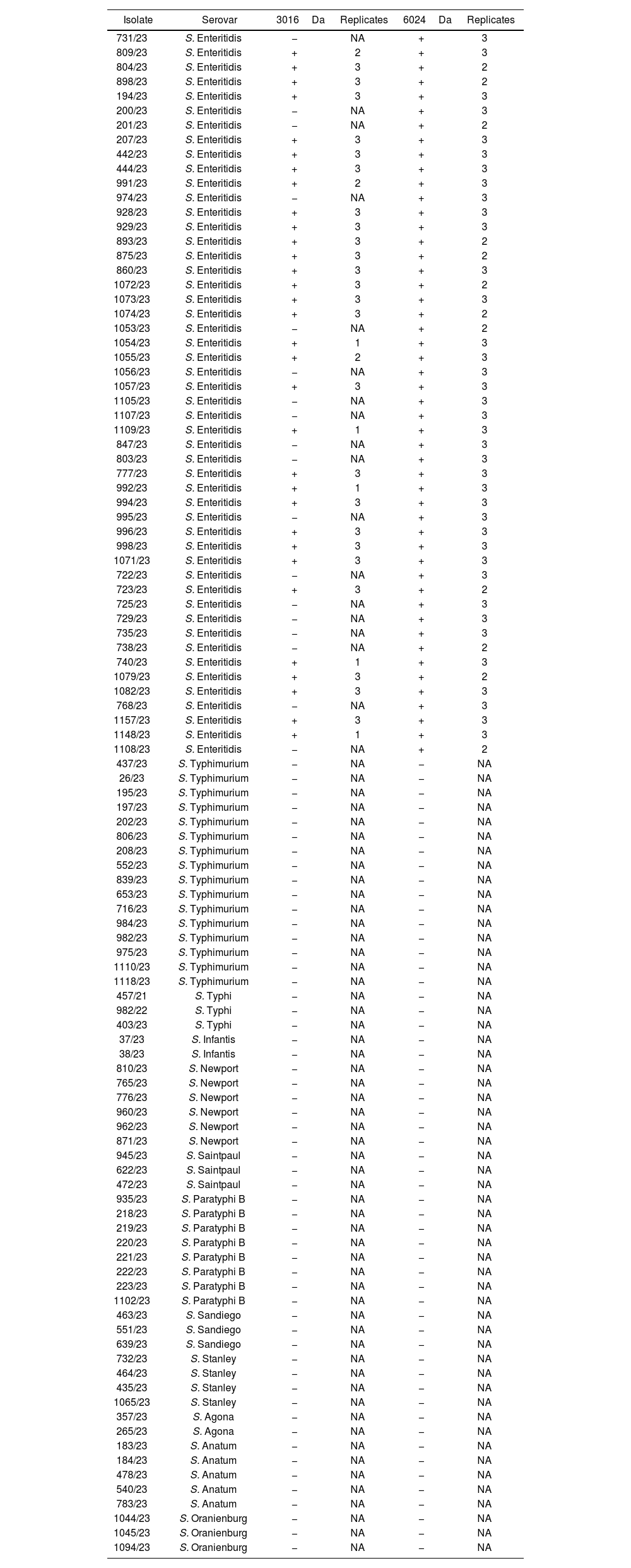
S. Enteritidis biomarkers visual detection with MALDI-TOF MS. The number of replicates in which the peaks were visually detected is indicated for each isolate.
| Isolate | Serovar | 3016Da | Replicates | 6024Da | Replicates |
|---|---|---|---|---|---|
| 731/23 | S. Enteritidis | − | NA | + | 3 |
| 809/23 | S. Enteritidis | + | 2 | + | 3 |
| 804/23 | S. Enteritidis | + | 3 | + | 2 |
| 898/23 | S. Enteritidis | + | 3 | + | 2 |
| 194/23 | S. Enteritidis | + | 3 | + | 3 |
| 200/23 | S. Enteritidis | − | NA | + | 3 |
| 201/23 | S. Enteritidis | − | NA | + | 2 |
| 207/23 | S. Enteritidis | + | 3 | + | 3 |
| 442/23 | S. Enteritidis | + | 3 | + | 3 |
| 444/23 | S. Enteritidis | + | 3 | + | 3 |
| 991/23 | S. Enteritidis | + | 2 | + | 3 |
| 974/23 | S. Enteritidis | − | NA | + | 3 |
| 928/23 | S. Enteritidis | + | 3 | + | 3 |
| 929/23 | S. Enteritidis | + | 3 | + | 3 |
| 893/23 | S. Enteritidis | + | 3 | + | 2 |
| 875/23 | S. Enteritidis | + | 3 | + | 2 |
| 860/23 | S. Enteritidis | + | 3 | + | 3 |
| 1072/23 | S. Enteritidis | + | 3 | + | 2 |
| 1073/23 | S. Enteritidis | + | 3 | + | 3 |
| 1074/23 | S. Enteritidis | + | 3 | + | 2 |
| 1053/23 | S. Enteritidis | − | NA | + | 2 |
| 1054/23 | S. Enteritidis | + | 1 | + | 3 |
| 1055/23 | S. Enteritidis | + | 2 | + | 3 |
| 1056/23 | S. Enteritidis | − | NA | + | 3 |
| 1057/23 | S. Enteritidis | + | 3 | + | 3 |
| 1105/23 | S. Enteritidis | − | NA | + | 3 |
| 1107/23 | S. Enteritidis | − | NA | + | 3 |
| 1109/23 | S. Enteritidis | + | 1 | + | 3 |
| 847/23 | S. Enteritidis | − | NA | + | 3 |
| 803/23 | S. Enteritidis | − | NA | + | 3 |
| 777/23 | S. Enteritidis | + | 3 | + | 3 |
| 992/23 | S. Enteritidis | + | 1 | + | 3 |
| 994/23 | S. Enteritidis | + | 3 | + | 3 |
| 995/23 | S. Enteritidis | − | NA | + | 3 |
| 996/23 | S. Enteritidis | + | 3 | + | 3 |
| 998/23 | S. Enteritidis | + | 3 | + | 3 |
| 1071/23 | S. Enteritidis | + | 3 | + | 3 |
| 722/23 | S. Enteritidis | − | NA | + | 3 |
| 723/23 | S. Enteritidis | + | 3 | + | 2 |
| 725/23 | S. Enteritidis | − | NA | + | 3 |
| 729/23 | S. Enteritidis | − | NA | + | 3 |
| 735/23 | S. Enteritidis | − | NA | + | 3 |
| 738/23 | S. Enteritidis | − | NA | + | 2 |
| 740/23 | S. Enteritidis | + | 1 | + | 3 |
| 1079/23 | S. Enteritidis | + | 3 | + | 2 |
| 1082/23 | S. Enteritidis | + | 3 | + | 3 |
| 768/23 | S. Enteritidis | − | NA | + | 3 |
| 1157/23 | S. Enteritidis | + | 3 | + | 3 |
| 1148/23 | S. Enteritidis | + | 1 | + | 3 |
| 1108/23 | S. Enteritidis | − | NA | + | 2 |
| 437/23 | S. Typhimurium | − | NA | − | NA |
| 26/23 | S. Typhimurium | − | NA | − | NA |
| 195/23 | S. Typhimurium | − | NA | − | NA |
| 197/23 | S. Typhimurium | − | NA | − | NA |
| 202/23 | S. Typhimurium | − | NA | − | NA |
| 806/23 | S. Typhimurium | − | NA | − | NA |
| 208/23 | S. Typhimurium | − | NA | − | NA |
| 552/23 | S. Typhimurium | − | NA | − | NA |
| 839/23 | S. Typhimurium | − | NA | − | NA |
| 653/23 | S. Typhimurium | − | NA | − | NA |
| 716/23 | S. Typhimurium | − | NA | − | NA |
| 984/23 | S. Typhimurium | − | NA | − | NA |
| 982/23 | S. Typhimurium | − | NA | − | NA |
| 975/23 | S. Typhimurium | − | NA | − | NA |
| 1110/23 | S. Typhimurium | − | NA | − | NA |
| 1118/23 | S. Typhimurium | − | NA | − | NA |
| 457/21 | S. Typhi | − | NA | − | NA |
| 982/22 | S. Typhi | − | NA | − | NA |
| 403/23 | S. Typhi | − | NA | − | NA |
| 37/23 | S. Infantis | − | NA | − | NA |
| 38/23 | S. Infantis | − | NA | − | NA |
| 810/23 | S. Newport | − | NA | − | NA |
| 765/23 | S. Newport | − | NA | − | NA |
| 776/23 | S. Newport | − | NA | − | NA |
| 960/23 | S. Newport | − | NA | − | NA |
| 962/23 | S. Newport | − | NA | − | NA |
| 871/23 | S. Newport | − | NA | − | NA |
| 945/23 | S. Saintpaul | − | NA | − | NA |
| 622/23 | S. Saintpaul | − | NA | − | NA |
| 472/23 | S. Saintpaul | − | NA | − | NA |
| 935/23 | S. Paratyphi B | − | NA | − | NA |
| 218/23 | S. Paratyphi B | − | NA | − | NA |
| 219/23 | S. Paratyphi B | − | NA | − | NA |
| 220/23 | S. Paratyphi B | − | NA | − | NA |
| 221/23 | S. Paratyphi B | − | NA | − | NA |
| 222/23 | S. Paratyphi B | − | NA | − | NA |
| 223/23 | S. Paratyphi B | − | NA | − | NA |
| 1102/23 | S. Paratyphi B | − | NA | − | NA |
| 463/23 | S. Sandiego | − | NA | − | NA |
| 551/23 | S. Sandiego | − | NA | − | NA |
| 639/23 | S. Sandiego | − | NA | − | NA |
| 732/23 | S. Stanley | − | NA | − | NA |
| 464/23 | S. Stanley | − | NA | − | NA |
| 435/23 | S. Stanley | − | NA | − | NA |
| 1065/23 | S. Stanley | − | NA | − | NA |
| 357/23 | S. Agona | − | NA | − | NA |
| 265/23 | S. Agona | − | NA | − | NA |
| 183/23 | S. Anatum | − | NA | − | NA |
| 184/23 | S. Anatum | − | NA | − | NA |
| 478/23 | S. Anatum | − | NA | − | NA |
| 540/23 | S. Anatum | − | NA | − | NA |
| 783/23 | S. Anatum | − | NA | − | NA |
| 1044/23 | S. Oranienburg | − | NA | − | NA |
| 1045/23 | S. Oranienburg | − | NA | − | NA |
| 1094/23 | S. Oranienburg | − | NA | − | NA |
+: positive (presence); −: negative (absence); NA: not applicable.
Both biomarkers were also detected visually after protein extraction with organic solvents (extraction method). In this case, peaks intensities were generally higher (255 a.u.–17500 a.u. for the ∼3016Da peak; 542 a.u.–45800 a.u. for the ∼6034Da peak) than those observed when performing the direct method.
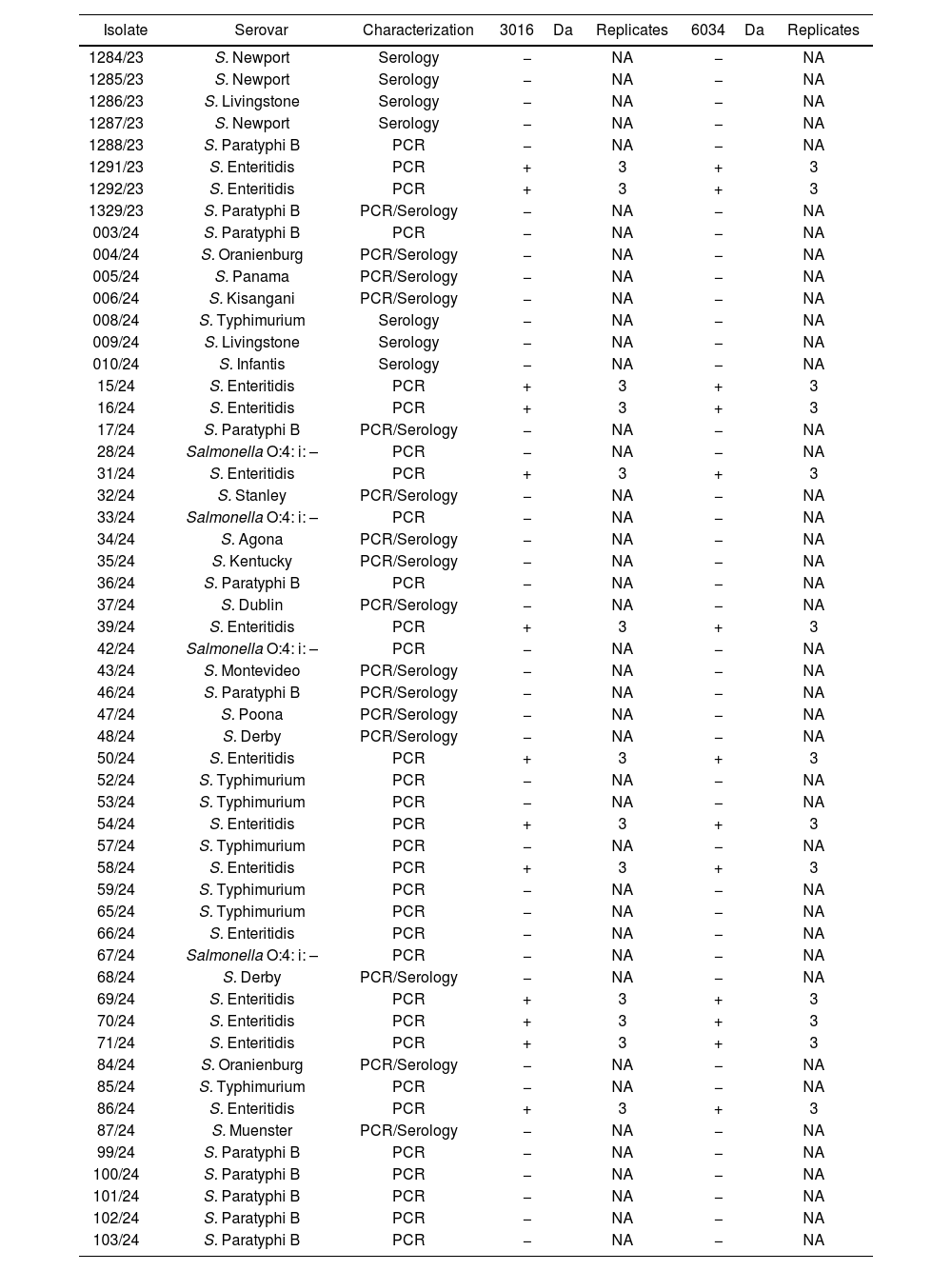
Biomarker blind search in Salmonella spp. isolatesS. Enteritidis serovar represented 25% of the strain collection. Concordance for S. Enteritidis identification between biomarker detection and serology/PCR assays performed afterwards was 98% (only one isolate −66/24− was identified as S. Enteritidis by PCR, and the specific biomarkers were not observed on the corresponding spectra). We observed both biomarkers (3016±3Da and 6034±3Da) in 13/55 Salmonella spp. isolates (Table 2).
Salmonella spp. isolates evaluated for blind biomarkers search.
| Isolate | Serovar | Characterization | 3016Da | Replicates | 6034Da | Replicates |
|---|---|---|---|---|---|---|
| 1284/23 | S. Newport | Serology | − | NA | − | NA |
| 1285/23 | S. Newport | Serology | − | NA | − | NA |
| 1286/23 | S. Livingstone | Serology | − | NA | − | NA |
| 1287/23 | S. Newport | Serology | − | NA | − | NA |
| 1288/23 | S. Paratyphi B | PCR | − | NA | − | NA |
| 1291/23 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 1292/23 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 1329/23 | S. Paratyphi B | PCR/Serology | − | NA | − | NA |
| 003/24 | S. Paratyphi B | PCR | − | NA | − | NA |
| 004/24 | S. Oranienburg | PCR/Serology | − | NA | − | NA |
| 005/24 | S. Panama | PCR/Serology | − | NA | − | NA |
| 006/24 | S. Kisangani | PCR/Serology | − | NA | − | NA |
| 008/24 | S. Typhimurium | Serology | − | NA | − | NA |
| 009/24 | S. Livingstone | Serology | − | NA | − | NA |
| 010/24 | S. Infantis | Serology | − | NA | − | NA |
| 15/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 16/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 17/24 | S. Paratyphi B | PCR/Serology | − | NA | − | NA |
| 28/24 | Salmonella O:4: i: – | PCR | − | NA | − | NA |
| 31/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 32/24 | S. Stanley | PCR/Serology | − | NA | − | NA |
| 33/24 | Salmonella O:4: i: – | PCR | − | NA | − | NA |
| 34/24 | S. Agona | PCR/Serology | − | NA | − | NA |
| 35/24 | S. Kentucky | PCR/Serology | − | NA | − | NA |
| 36/24 | S. Paratyphi B | PCR | − | NA | − | NA |
| 37/24 | S. Dublin | PCR/Serology | − | NA | − | NA |
| 39/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 42/24 | Salmonella O:4: i: – | PCR | − | NA | − | NA |
| 43/24 | S. Montevideo | PCR/Serology | − | NA | − | NA |
| 46/24 | S. Paratyphi B | PCR/Serology | − | NA | − | NA |
| 47/24 | S. Poona | PCR/Serology | − | NA | − | NA |
| 48/24 | S. Derby | PCR/Serology | − | NA | − | NA |
| 50/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 52/24 | S. Typhimurium | PCR | − | NA | − | NA |
| 53/24 | S. Typhimurium | PCR | − | NA | − | NA |
| 54/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 57/24 | S. Typhimurium | PCR | − | NA | − | NA |
| 58/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 59/24 | S. Typhimurium | PCR | − | NA | − | NA |
| 65/24 | S. Typhimurium | PCR | − | NA | − | NA |
| 66/24 | S. Enteritidis | PCR | − | NA | − | NA |
| 67/24 | Salmonella O:4: i: – | PCR | − | NA | − | NA |
| 68/24 | S. Derby | PCR/Serology | − | NA | − | NA |
| 69/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 70/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 71/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 84/24 | S. Oranienburg | PCR/Serology | − | NA | − | NA |
| 85/24 | S. Typhimurium | PCR | − | NA | − | NA |
| 86/24 | S. Enteritidis | PCR | + | 3 | + | 3 |
| 87/24 | S. Muenster | PCR/Serology | − | NA | − | NA |
| 99/24 | S. Paratyphi B | PCR | − | NA | − | NA |
| 100/24 | S. Paratyphi B | PCR | − | NA | − | NA |
| 101/24 | S. Paratyphi B | PCR | − | NA | − | NA |
| 102/24 | S. Paratyphi B | PCR | − | NA | − | NA |
| 103/24 | S. Paratyphi B | PCR | − | NA | − | NA |
+: positive (presence); −: negative (absence); NA: not applicable.
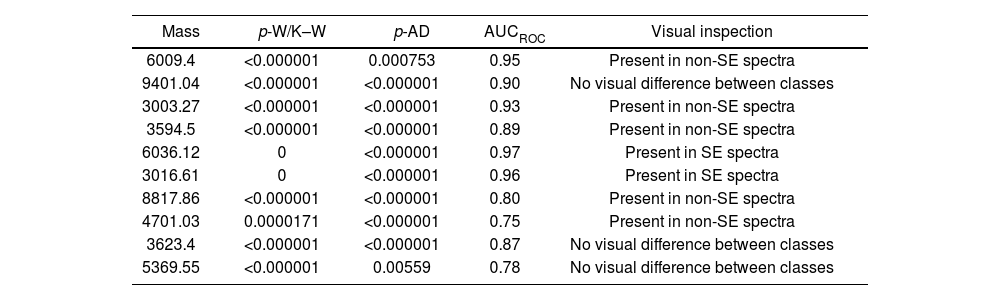
Statistical analysis of spectra acquired for blind biomarker detection showed a p-value <0.05 for the AD and W/K–W tests for the best ten discriminating class peaks, indicating a significant difference for the selected biomarkers (3016.61Da and 6036.12Da peaks) when S. Enteritidis and non-S. Enteritidis groups were evaluated (Table 3). This mass list included the biomarkers previously evaluated visually for S. Enteritidis detection. The AUCROC was 0.96 and 0.97 for the 3016.61Da and 6036.12Da selected peaks, respectively. Some biomarkers selected by the ClinPro Tools software were detected in non-S. Enteritidis spectra, also showing high AUCROC values (Table 3).
Top ten differentiating peaks selected by ClinPro Tools v S. 3.0 for S. Enteritidis and non-S. Enteritidis spectra.
| Mass | p-W/K–W | p-AD | AUCROC | Visual inspection |
|---|---|---|---|---|
| 6009.4 | <0.000001 | 0.000753 | 0.95 | Present in non-SE spectra |
| 9401.04 | <0.000001 | <0.000001 | 0.90 | No visual difference between classes |
| 3003.27 | <0.000001 | <0.000001 | 0.93 | Present in non-SE spectra |
| 3594.5 | <0.000001 | <0.000001 | 0.89 | Present in non-SE spectra |
| 6036.12 | 0 | <0.000001 | 0.97 | Present in SE spectra |
| 3016.61 | 0 | <0.000001 | 0.96 | Present in SE spectra |
| 8817.86 | <0.000001 | <0.000001 | 0.80 | Present in non-SE spectra |
| 4701.03 | 0.0000171 | <0.000001 | 0.75 | Present in non-SE spectra |
| 3623.4 | <0.000001 | <0.000001 | 0.87 | No visual difference between classes |
| 5369.55 | <0.000001 | 0.00559 | 0.78 | No visual difference between classes |
W/K–W: Wilcoxon/Kruskal–Wallis test; AD: Anderson–Darling test; SE: S. Enteritidis.
In this study, we analyzed the utility of MALDI-TOF MS to detect previously described S. Enteritidis specific biomarkers18, using local strains. The protocol is simple and fast, and is the same as the one used for bacterial identification with MALDI-TOF MS, so after achieving a “Salmonella spp.” result by the automated identification protocol, visual inspection of the same spectra could be performed for biomarker searching.
The investigated biomarkers were detected from TSA media, and peak visualization was not possible from SS Agar, probably due to the presence of inhibitors. Other solid culture media such as XLD or MacConkey Agar usually employed for Salmonella isolation in the clinical setting could be tested.
We observed variable intensities for the selected biomarkers in the retrospective study, and we recommend the evaluation of the signal-to-noise ratio of every single spectrum, before arriving to a concluding report. In addition, both biomarkers were detected when performing protein extraction with organic solvents in some samples.
Blind biomarker search in Salmonella spp. isolates displayed a perfect concordance with the serovar results obtained by methods daily employed in our laboratory. Statistical analysis with ClinPro Tools software supported the results obtained by visual analysis, with high AUCROC values for the evaluated biomarkers, indicating a great discriminative power between classes12. Detection from conserved isolates (retrospective study) showed a poorer performance than from fresh isolates (blind search), probably due to different microorganism storage lapses.
Visual detection of these S. Enteritidis biomarkers could be used as a screening method for serovars differentiation, after Salmonella spp. identification is achieved. We consider that S. Enteritidis should be reported as “presumptive” or “preliminary” with this methodology, as it was only possible to analyze a limited, though locally representative, set of Salmonella serovars. A more extensive analysis would be necessary, including as many serovars as possible, to make more concluding observations. Another limitation, in addition to the isolation on culture medium (TSA), limited report (“presumptive” or “preliminary” result) and limited serovars evaluated, would be that laboratory technicians should be trained for biomarker visual detection with the available software.
ConclusionsMALDI-TOF MS technology can be used for presumptive S. Enteritidis differentiation from other serovars, by serovar-specific biomarkers detection, reducing TAT results compared with serology and genomic approaches. This technology has proven to be useful for various applications, with rapid subtyping of microorganisms being one of the approaches that still needs to be intensified and can be advantageous for making therapeutic decisions in the clinical setting. Furthermore, it would be useful to provide a rapid response to take action and control this serovars of Salmonella associated mainly with food outbreaks in the country.
FundingThis study was supported by Instituto Nacional de Enfermedades Infecciosas, INEI-ANLIS-“Carlos G. Malbrán”, Buenos Aires, Argentina.
Conflict of interestThe authors declare no commercial or financial conflict of interest.
We deeply thank María Florencia Rocca and Mónica Prieto, from Bacteriología Especial Service (ANLIS-Malbrán), for their technical and scientific support, and researchers from Unidad Operativa Centro Nacional de Genómica y Bioinformática (UOCNGB-ANLIS-Malbrán) for isolates sequencing.