The confirmation of two cases of hantavirus pulmonary syndrome (HPS) in the city of San Miguel de Tucuman, province of Tucuman, is described in the present study. In 2021, the diagnosis of two patients with HPS with no history of travel outside the province was confirmed. The infection was determined by serology in both cases. Case 1 had an unfavorable outcome, was confirmed by the detection of specific IgM antibodies for orthohantavirus and the identification in a serum sample from three days of evolution of a viral genotype, which was previously unreported to be in circulation in Argentina. In Case 2, specific IgM and IgG type antibodies were detected in two serum samples, with no amplification of the viral genome. Due to the recreational habits of the patients, it is possible to infer exposure to infected rodent fluids during outdoor activities in their areas of residence. In conclusion, the circulation of a new viral genotype of orthohantavirus in Argentina is confirmed.
Se describe la confirmación de dos casos del síndrome pulmonar por hantavirus (SPH) en la ciudad de San Miguel de Tucumán, provincia de Tucumán. En el año 2021, se confirmó mediante serología el diagnóstico de SPH en dos personas sin antecedentes de viaje fuera de la provincia. Uno de los casos (caso 1), que tuvo una evolución desfavorable, se confirmó mediante la detección de IgM específica de orthohantavirus; se trató de un genotipo viral desconocido en Argentina en una muestra de suero de 3 días de evolución. En el otro individuo (caso 2), se detectaron anticuerpos específicos tipo IgM e IgG en dos muestras de suero sin amplificación de genoma viral. Debido a los hábitos recreativos de los pacientes, es posible inferir la exposición a fluidos de roedores infectados durante actividades recreativas al aire libre en sus áreas de residencia. En conclusión, se demuestra la circulación de un nuevo genotipo viral de orthohantavirus en Argentina.
Hantavirus pulmonary syndrome (HPS) is an acute viral disease caused by members of the genus Orthohantavirus, family Hantaviridae12. The main route of transmission to humans is through aerosolization in the environment of excreta from infected rodents. However, Andes virus is the only orthohantavirus with evidence of person-to-person transmission, generating a public health concern due to its high mortality rate (25–35%)15.
Currently, The International Committee on Taxonomy of Viruses (ICTV) recognizes two species of orthohantavirus associated with HPS circulating in Argentina: Orthohantavirus andesense and Orthohantavirus negraense6. The O. andesense species is represented by Andes, Lechiguanas and Orán viruses; while the O. negraense species is represented by Laguna Negra virus. There are other orthohantavirus genotypes circulating in Argentina that do not meet the criteria for classification into the recognized orthohantavirus species: Juquitiba, Bermejo, HU39694 and Alto Paraguay genotypes are associated with HPS and Maciel, Pergamino, Leyes and Seoul genotypes have not yet been associated with disease in humans2,9,11,13.
Before 2016, Tucuman was not considered a potential endemic area for hantavirus, despite the identification of hantavirus rodent reservoirs in the province14,5,16.
Since 2016, three confirmed cases with no history of travel outside the province were identified. Two cases in 2016, with probable exposure in the department of Burruyacú, both deceased, and one case in 2017, with probable exposure in the department of Leales, with favorable outcome. Phylogenetic studies allowed identification of the HU39694 genotype4.
The aim of the present work was to describe the first clinical case of HPS caused by an unknown genotype of orthohantavirus in the province of Tucuman, Argentina.
Case 1. A previously healthy young woman, residing in San Miguel de Tucuman, attended a health care center presenting with a 5-day history of fever and asthenia. At the time of admission, on October 23, 2021, she had an acute respiratory illness, with progression to respiratory failure, needing hospitalization in an intensive care unit and mechanical ventilation. Laboratory results revealed a reduction in platelet count, high hematocrit and lactic acidosis. She exhibited active bleeding in brain and lungs, and had a fatal outcome within hours of admission. On October 21, the patient had undergone a nasopharyngeal swab, and the sample tested negative for SARS-CoV-2 using an antigen test. On October 22, a second sample was processed by the RT-PCR technique with non-detectable result. She had visited rural areas, which were considered potential exposure sites.
Case 2. A young healthy male from San Miguel de Tucuman attended a health care institution with a 4-day history of fever, nausea, headache and abdominal pain on October 24, 2021. He had a diagnosis of right basal pneumonia, received empirical antibiotic treatment and required high-flow cannula for 2 days. COVID-19, dengue, leptospirosis and bacterial infection due to common germs were ruled out. Due to his favorable evolution, he was discharged 9 days after admission. Twenty-five days before he had noticed the initial symptoms, after camping in a mountain area for three days.
Serum samples from Case 1 and Case 2 patients (two serum samples collected on different days of evolution, 9 and 18 days) were received at the Instituto Nacional de Enfermedades Virales Humanas (INEVH-ANLIS), Pergamino, Argentina, for HPS laboratory diagnosis. The search for antibodies was carried out using the ELISA technique for the detection of immunoglobulin M (IgM) and immunoglobulin G (IgG), which are specific for orthohantavirus using the Maciel genotype antigen4.
For the detection of the viral genome, the total RNA of the patient's serum samples was extracted using with the Qiamp kit Viral RNA Mini Kit (Qiagen Inc., Germany), following the manufacturer's recommendations. The amplification of the genome was performed by nested RT-PCR, using generic oligonucleotides for orthohantaviruses that amplify a 447 nt fragment of the nucleoprotein (NP) (S segment) and a 513 nt fragment of G2 (M segment), based on previously published conditions4.
For the identification of the circulating genotype, the amplification products obtained were purified using the QIAquick Gel Extraction Kit (Qiagen Inc., Germany), following the manufacturer's recommendations, sequenced with the BigDye Terminator v3.1 Cycle Sequencing Kit in a 3500 Avant Genetic Analyzer (Applied Biosystems) and subjected to a global comparison analysis with BLAST to determine similarities with species of the genus Orthohantavirus1. Sequence alignment was constructed using Muscle (v3.8.425) in AliView (v1.28) software8. The determination of the evolutionary models was performed using the program ModelFinder7. Phylogenetic analyses of NP and G2 fragments were conducted by maximum likelihood with W-IQ-TREE18. Statistical support for clades was calculated using ultrafast bootstrap analysis, with the performance of 2000 replicates. Trees were visualized and edited using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/). Nucleotide similarity percentages were calculated using the MEGA11 program17.
HPS diagnostic confirmation of the cases was obtained through the detection of specific IgM type antibodies by ELISA, with a titer greater or equal to 6400 in the serum sample with 3 days of evolution for Case 1 and in the serum sample from 9 days of evolution for Case 2 since the onset of symptoms. No IgG type antibodies were detected in the acute sample for Case 1. For Case 2, a titer of 1600 by ELISA IgG was detected in the 9-day sample. In the second 18-day sample, the serological titers previously obtained for IgM and IgG were maintained.
Genomic fragments of the S and M segments of the viral genome were amplified from the Case 1 sample (GenBank no. PQ248942 for the S segment and PQ248943 for the M segment). The global comparison of the NP and G2 sequences using BLAST showed 82–87% identity with the known orthohantavirus genotypes available on GenBank, suggesting that this could be a new orthohantavirus genotype. Furthermore, they exhibited 92–96% identity with PQ133337, PQ133338, PQ133339 for the S segment and PQ156450, PQ156451 for the M segment. These sequences correspond to rodents of the genus Oligoryzomys captured in the province of Salta, Argentina in 2014 in the context of an HPS case, previously studied at INEVH but whose data have not been published.
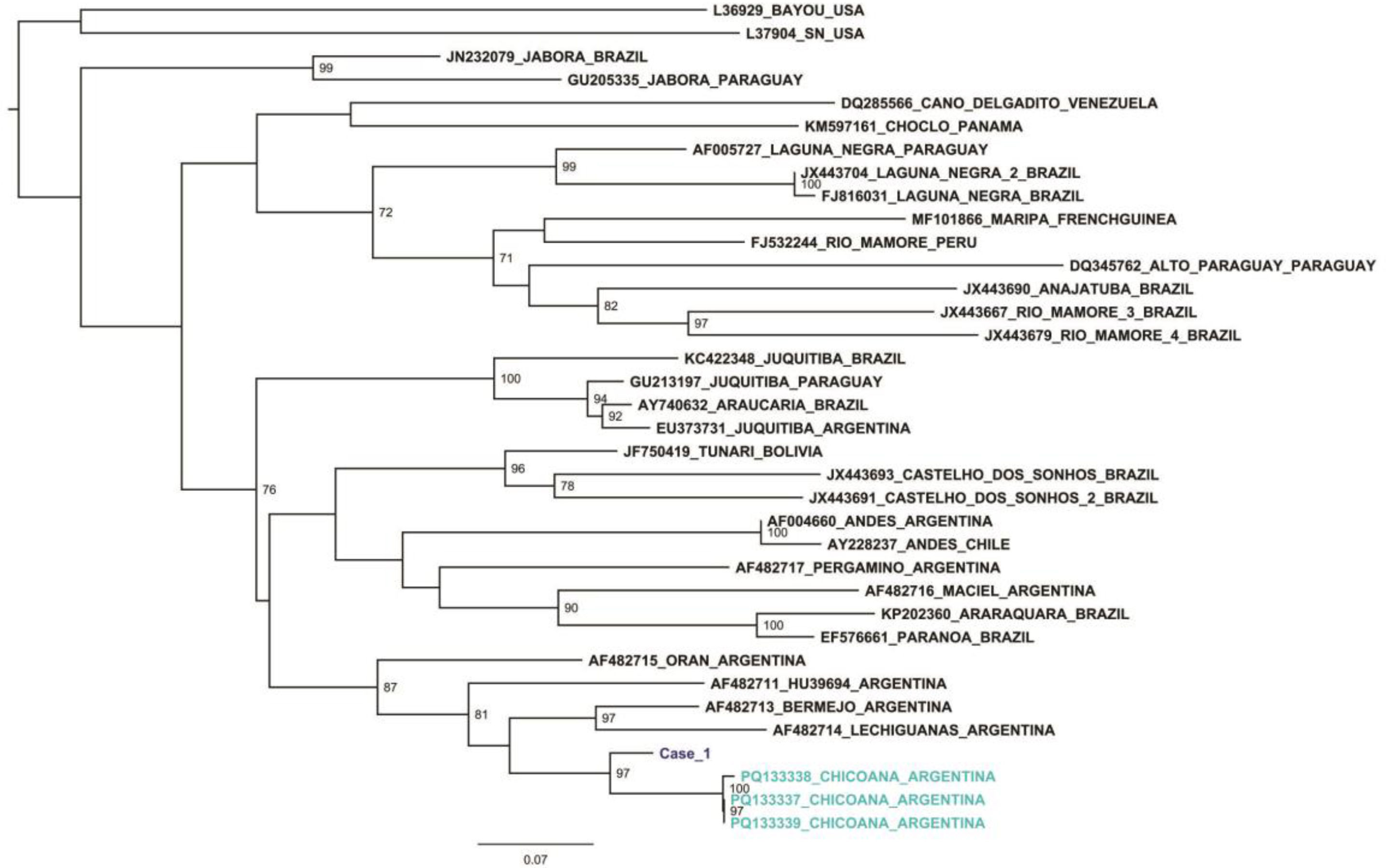
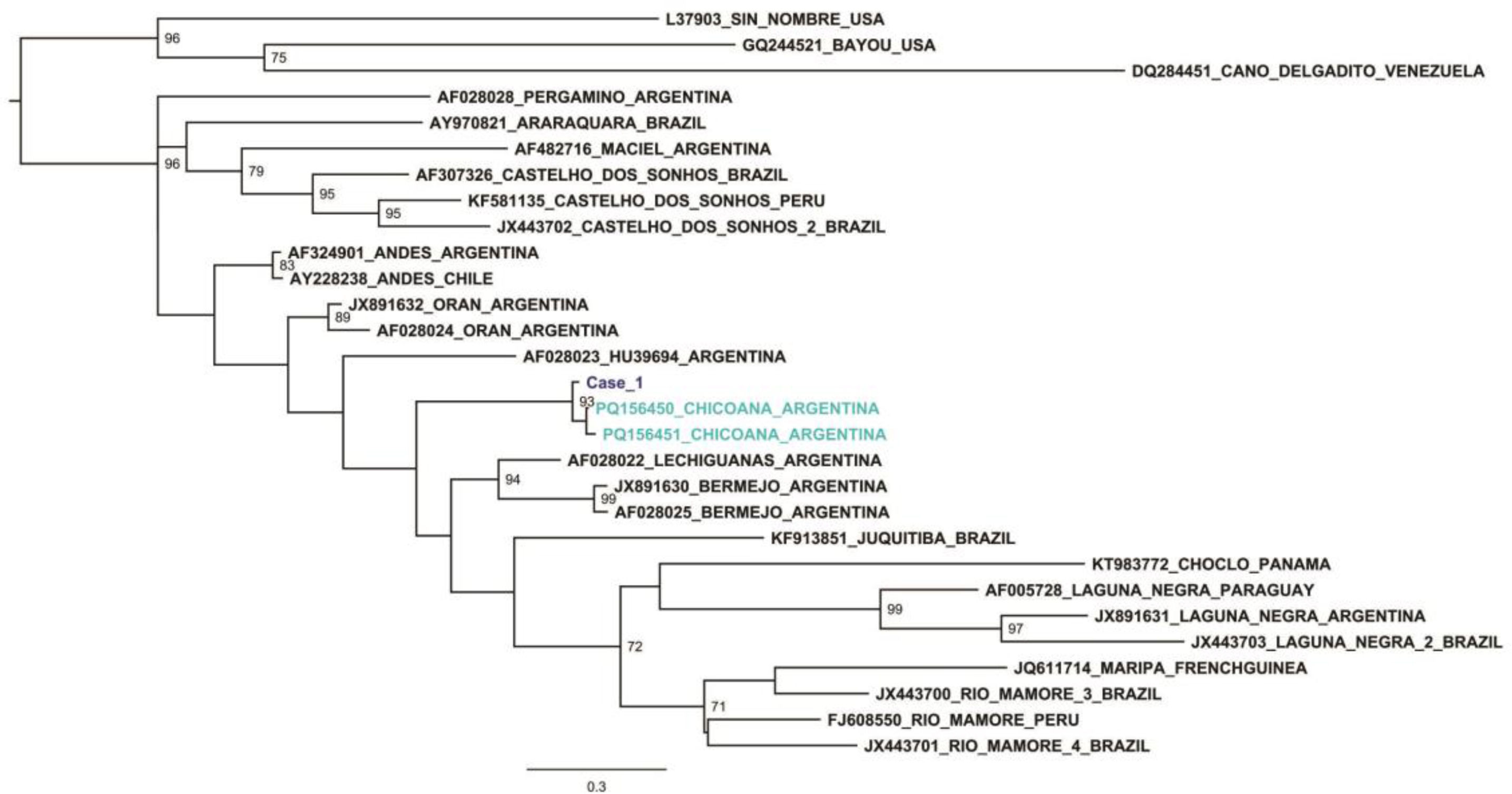
Figures 1 and 2 show the phylogenetic reconstructions obtained by maximum likelihood for a 428 nucleotide fragment of the NP (S segment) and a 329 nucleotide fragment for G2 (M segment), respectively. The sequences obtained from Case 1 grouped together with the sequences from the Oligoryzomys sp. from province of Salta, forming a well-supported clade. The bootstrap support values are detailed in the nodes of the phylogenetic trees. Comparison of the viral sequences obtained from the Case 1 sample with the sequences from the Oligoryzomys sp. from province of Salta showed 92.1–92.3% and 95.4–96.4% nucleotide similarity for the S segment and M segment, respectively. The comparison with other orthohantavirus sequences from America and Argentina used for the phylogenetic reconstructions showed 77–87.4% and 72.9–84.5% nucleotide similarity for the S and M segments, respectively.
Maximum likelihood tree for a 428 nt fragment of the NP (S segment). The tree was obtained with W-IQ-TREE using TIM2+F+I+G4 as model of nucleotide substitution for the S dataset. The sequences of published orthohantavirus genotypes were named according to GenBank accession number, circulating genotype and country of origin. The numbers in the nodes indicate statistical support values (only values greater than 70% are shown). The sequence from Case 1 is highlighted in dark blue.
Maximum likelihood tree for a 329 nt fragment of the G2 (M segment). The tree was obtained with W-IQ-TREE using GTR+F+I+G4 as model of nucleotide substitution for the M dataset. The sequences of published orthohantavirus genotypes were named according to GenBank accession number, circulating genotype and country of origin. The numbers in the nodes indicate statistical support values (only values greater than 70% are shown). The sequence from Case 1 is highlighted in dark blue.
The results of the present study provide strong evidence of the circulation of a potentially new pathogenic viral genotype in Argentina. When analyzing the nucleotide similarity between the sequences from Case 1 and known orthohantavirus genotypes from Argentina, 78.8–87.5% and 76.9–84.5% for the S and M segments respectively, could be observed, but a higher similarity was found between this sequence and the ones from the Oligoryzomys sp. from the province of Salta. When analyzing the intragenotype variability of orthohantaviruses circulating in Argentina, 92–99% of nucleotide identities is generally observed3,10. The relationship between these sequences suggest that they belong to the same orthohantavirus genotype (proposed name: Orthohantavirus chicoanense, proposed genotype: Chicoana) and are associated with HPS in the province of Tucuman. Complete genome sequencing will provide more information of this new genotype and its relationship with other orthohantavirus genotypes described so far. The presence of this new genotype in two provinces of northern Argentina suggests a wide circulation of this pathogenic genotype; hence, further ecoepidemiological studies are needed to confirm the proposed hypothesis and to detect the rodent reservoir. Accurate species identification and range mapping would enhance the understanding of the epidemiology of this disease.
FundingThis study was funded by ANLIS.
Conflict of interestThe authors declare no conflicts of interest.
To Natalia Calandri and Ayelén Barrio for technical support in serological diagnosis.