Hantavirus Pulmonary Syndrome (HPS) is an emerging infectious disease of the Americas. Eight native rodent species have been identified as HPS virus reservoirs in Argentina. The aim of this work was to detect the orthohantavirus genotypes present in a rodent community that inhabits a zone where a fatal HPS case occurred within an endemic locality of Central Argentina. We captured 27 rodents with a trapping effort of 723 trap nights. We detected 14.3% of infected Akodon azarae with the Pergamino genotype. This result expands the known distribution of this orthohantavirus. Although the Pergamino genotype has not been associated with human cases, the information about its distribution is relevant for risk assessment against potential changes in the virus infectivity.
El síndrome pulmonar por hantavirus (SPH) es una enfermedad infecciosa emergente en América. Ocho especies de roedores nativos han sido identificadas como reservorios del virus causante del SPH en la Argentina. El objetivo de este trabajo fue detectar los genotipos de orthohantavirus presentes en una comunidad de roedores que habita en una zona donde ocurrió un caso fatal de SPH, en una localidad endémica de Argentina central. Se capturaron 27 individuos con un esfuerzo de 723 trampas-noche. Se detectó un 14,3% de Akodon azarae infectados con el genotipo pergamino. Este resultado amplía el conocido rango de distribución de este orthohantavirus. A pesar de que el genotipo pergamino no ha sido asociado con casos humanos hasta el momento, la información sobre su distribución es relevante para analizar el riesgo ante un potencial cambio en la infectividad del virus.
Hantavirus Pulmonary Syndrome (HPS) is a zoonotic emerging infectious disease characterized by a rapid onset of pulmonary edema followed by respiratory failure and cardiogenic shock. It is caused by New World orthohantaviruses (family Hantaviridae, subfamily Mammantavirinae, genus Orthohantavirus) which are hosted by rodents of the family Cricetidae. In Argentina, since the characterization of the Andes virus (ANDV) in 19958, different variants or genotypes of this virus have been described. Eight native species have been identified as orthohantavirus hosts: Oligoryzomys flavescens, O. nigripes, O. longicaudatus, O. chacoensis, O. occidentalis, Calomys callosus, Necromys obscurus and Akodonazarae4,6,7,14,17,21. Individuals of other species, such as Abrothrix hirta, A. olivacea, Loxodontomys micropus, Deltamys kempi and Oxymycterus rufus have occasionally been found to be seropositive, though these cases are often considered the result of spillover events3,15. Three orthohantavirus hosts are known to be present in Central-East Argentina: O. flavescens, host of Lechiguanas (LECV), Buenos Aires (BASV) and Plata (PTV) genotypes in Buenos Aires4,14 and Entre Ríos (only LECV)21; Necromys obscurus, host of Maciel (MACV) genotype in Buenos Aires; and A. azarae, host of Pergamino (PERV) genotype in Buenos Aires14. All these genotypes cause HPS with the exception of the PERV and MACV that have not yet been associated with human cases14.
The PERV genotype was detected for the first time in A. azarae tissues from Buenos Aires, Argentina7 and infected individuals were detected in subsequent investigations carried out in the same province9,21. However, all the studies conducted in other neighboring provinces such as Entre Rios and Santa Fe did not detect infected A. azarae despite the great capture effort9,19,21.
Due to the fact that there is no specific treatment available for HPS, prevention is the only possible measure to reduce the morbidity and mortality caused by this disease. The identification of the reservoir hosts and the delimitation of both the geographical range of the host and that of the virus within the host range are crucial parts of the knowledge needed to design effective preventive measures20. The aim of this work was to detect the orthohantavirus genotypes present in a rodent community that inhabits a zone where a fatal HPS case occurred within an endemic locality of Central Argentina.
This study was performed in a rural area of the Gualeguaychú department in Entre Ríos province, Argentina (33°03′53.1″S 58°52′39.1″W), which is located in Central-East Argentina, the second region in Argentina in terms of the number of HPS cases1,10. Based on a fatal HPS case that occurred in January 2018 the provincial health authorities requested the study of rodent assembles. In October 2018, small rodents were captured with live-traps baited with a mixture of fat, oats and peanut butter. Two hundred and forty one traps were located at 10m intervals during three consecutive nights on a poultry farm where the HPS patient used to work(composed of five breeding sheds and two sheds for storing tools and animal feed), in the patient's dwelling (with breeding animals such as pigs in the peridomicile) and near the vegetated margins of the stream where the patient used to go fishing.
For each animal captured, we recorded the species according to external morphology, sex and body measurements. To detect hantavirus-specific immunoglobulin G (IgG) antibodies, enzyme-linked immunosorbent assays were performed on blood samples13. We used antibody presence as an indication of infection. The animals were sacrificed and the tissues were stored in a nitrogen tank to determine the orthohantavirus genotype. All animals were handled according to the Argentine law for the protection of animal welfare (Penal Code Law No. 14346) and safety precautions appropriate for handling the orthohantavirus reservoirs were followed.
Total RNA was extracted from the lung tissues of seropositive rodents using Trizol (Invitrogen) and purified by the RNAid kit (QbioGene). Partial S-segment was amplified by RT-PCR followed by a second round of nested PCR. Specific oligonucleotide primers, based on conserved regions of the ANDV virus genome, were used2. End-point PCR reactions were carried out in an Applied Biosystems 2720 Thermal Cycler. For the S segment, the targets were a fragment of 840 nt (position 86–926) in length, corresponding to the nucleocapsid protein coding region. The PCR products were separated by agarose gel electrophoresis, purified and sequenced in an automated sequencer ABI Prism R3500 Genetic Analyzer (Applied Biosystems, UK) using the ABI PRISM Big Dye Terminator Sequencing kit (Applied Biosystems, UK). The obtained sequences were analyzed and compared with published sequences using MEGA 10 software5.
Models were developed to describe substitution patterns and were selected by BIC (Bayesian Information Criterion). Models with the lowest BIC scores are considered to best describe the substitution pattern. For each model, AICc value (Akaike Information Criterion, corrected), Maximum Likelihood value (lnL), and the number of parameters (including branch lengths) are also estimated12. For estimating the ML values, a tree topology was automatically computed. The analysis involved 15 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated.
We captured a total of 27 individuals with a trapping effort of 723 trap nights. Akodon azarae was the most abundant species (14) followed by Oxymycterus rufus (7), Calomys callidus (5) and Holochilus sp. (1). The first two species were captured both on the farm and in the proximities of the stream, while Calomys callidus was captured only on the farm and Holochilus sp. only in the stream. No rodents were captured around the patient's dwelling.
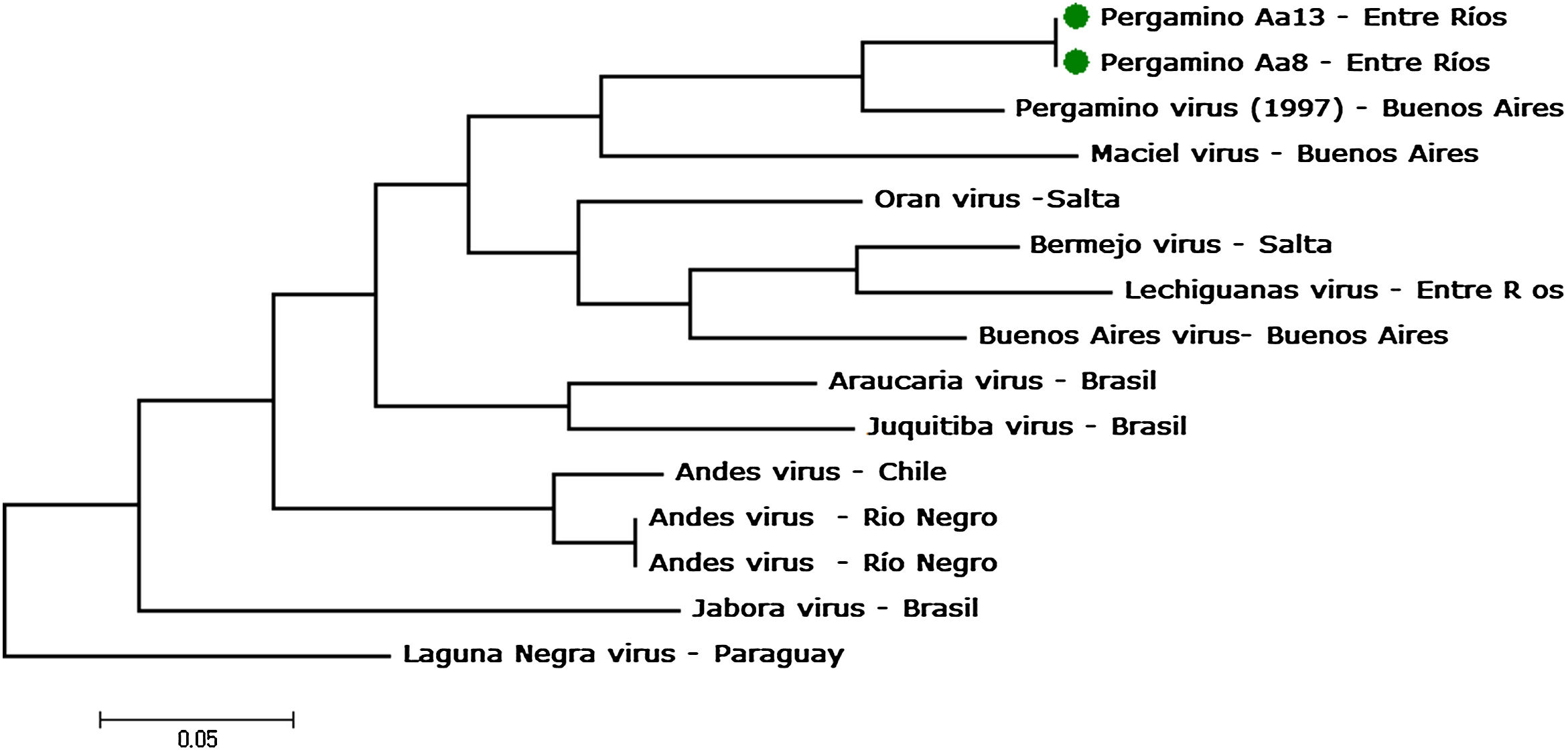
Two individuals of A. azarae (14.3%) were infected. Viral RNA was recovered from rodent tissues, partial fragments of S and M genomic segments were amplified, the nucleotide sequence determined and the phylogenetic analysis performed. Amplifications of all partial fragments were successful and the comparative analysis with other sequences was published in GENBANK, and in accordance with the criterion established by the ICTV and Rivera et al.16, showed the highest identities with the PERV genotype (strain 14403) for the S-segment fragment. The percentages of identity were 93% and 98% at nucleotide and amino acid level (accession numbers AF482717 and AAL82652, respectively, Fig. 1).
Molecular phylogenetic analysis by Maximum Likelihood method. Phylogenetic analysis of hantaviruses based on 853bp of S-segment genome of Andes virus, and other orthohantaviruses characterized previously. Taxa in the branches of the tree indicate virus name and province of origin. GenBank accession numbers and reservoir rodent specie were: AF482717, Pergamino (Akodon azarae); AF482716, Maciel (Necromys benefactus); AF482715, Orán (Oligoryzomys chacoensis); AF482713, Bermejo (Oligoryzomys occidentalis); AF482714, Lechiguanas (Oligoryzomysflavescens and O. nigripes); AF482711, Buenos Aires (undescribed); AY740624, Araucaria (O. nigripes); KC422346, Juquitiba (O. nigripes); AF291702, AF004660 and AF324902 Andes (Oligoryzomys longicaudatus; HM582091, Jaborá (Akodon montensis) and NC038505 Laguna Negra (Calomys laucha). The model selection retained GTR+G+I as the best model that shows the lowest Akaike (6384.96) and BIC (6645.48) values.
Despite not finding a pathogenic genotype that could be associated with the fatal HPS case recorded, the principal success of this research was to detect, for the first time, infected Akodon azarae with the PERV genotype in Entre Rios province. Although this genotype has not been associated with human cases, the information about its distribution could be relevant to assess risk in the case of changes in the infectivity of the virus11, as was the case with the Tula virus, the pathogenic agent of hemorrhagic fever with renal syndrome18. Additionally, the LECV genotype (associated with O. flavescens) was detected in O. nigripes of Entre Rios21 and Buenos Aires3 and PERV genotype (associated with A.azarae) in Oxymycterusrufus in Buenos Aires3, two non typical rodent species hosts, suggesting that host switching is frequent in this area. Due to these facts, permanent surveillance would be essential for the detection of the expansion of both reservoir host ranges and viral distribution in the reservoir population, as well as other factors associated with anthropogenic changes. These conditions should favor a potential impact on public health. Emerging infections are diseases that newly appear in a population, or existed previously but emerge due to dynamic interactions between rapidly evolving infectious agents and changes in the environment, such as perturbations, and changes in host's behavior that favor new ecological niches11. This first record of the PERV genotype in Entre Ríos expands the virus distribution known until now and complements the epidemiological monitoring and prevention of HPS in the province.
FundingThis research was funded by Zoonosis and Vectors Program of the Entre Ríos Health Ministry, University of Buenos Aires Q3 and CONICET.
Authors’ contributionsIEGV and JGA designed the research; IEGV, JGA and EFB did the fieldwork; IEGV and EFB write of the manuscript; RMC, CMB and VPM did the serological and molecular analyses; IEGV, JGA and VPM funded the research.
Conflict of interestThe authors declare that they have no conflicts of interest.
We thank to the personnel and authorities of the Entre Ríos Health Ministry for their logistical support.