Escherichia coli ETEC, EPEC, NTEC and STEC/EHEC pathotypes are often isolated from bovine feces. The objective of this study was to detect 21 E. coli virulence genes in feces from 252 dairy calves in Uruguay (149 with neonatal diarrhea – NCD – and 103 asymptomatic). Genes iucD, f17A, afa8E, papC, clpG and f17G(II) were the most prevalent (81.3%; 48.4%; 37.3%; 35.7%; 34.1%; 31.3%, respectively). Genes eae, stx1and stx2 were poorly represented; 13/252 animals harbored one or a combination of these genes. The prevalence of the cnf gene was 4.4%, while that of cdt-IV and cdt-III genes was 24.2% and 12.7% respectively. This study reports updated data about the virulence profiles of E. coli in dairy calves in Uruguay. A large number of adhesins and toxin genes were detected. Our results demonstrate that E. coli from bovine feces has diarrheagenic and extraintestinal profiles although other NCD risks factors may contribute to the disease outcome.
Los patotipos de Escherichia coli ETEC, EPEC, NTEC y STEC/EHEC son frecuentemente aislados de heces bovinas. El objetivo del presente estudio fue detectar 21 genes de virulencia de E. coli en las heces de 252 terneros de leche en Uruguay, 149 de ellos con síntomas de diarrea neonatal (DNT) y 103 asintomáticos. Los genes iucD, f17A, afa8E, papC, clpG y f17G(II) fueron los más prevalentes (81,3; 48,4; 37,3; 35,7; 34,1 y 31,3%, respectivamente). Los genes eae, stx1 y stx2 estuvieron poco representados: 13/252 animales presentaron uno o una combinación de dichos genes. La prevalencia del gen cnf fue del 4,4%, mientras que la de los genes cdt-IV y cdt-III fue del 24,2 y 12,7%, respectivamente. Este trabajo aporta datos actualizados sobre el perfil de virulencia de E. coli en terneros en Uruguay. Fueron detectados un alto número de genes de adherencia y de toxinas. Se demuestra que los aislamientos de E. coli recuperados de heces de terneros presentan perfiles diarreogénicos y extraintestinales, aunque otros factores de riesgo de DNT podrán contribuir al desarrollo de la enfermedad.
Escherichia coli is a widely distributed gram-negative bacterium, and is the most numerous facultative anaerobe inhabiting the intestine of warm-blooded animals2. In this niche, these bacteria coexist with other microorganisms assembling the commensal gut microbiota. However, some E. coli variants have acquired specific attributes that allow them to infect the immunocompetent host and cause disease4.
Pathogenic E. coli comprises a vast number of virulent variants associated with human and animal illnesses. Such is the diversity of virulence factors, together with the ability of bacteria to horizontally transfer several virulence-related genes through plasmids, phages and transposons, that nowadays at least 9 different pathotypes or virulent variants have been described4,6. Enterotoxigenic E. coli (ETEC) is characterized by the expression of fimbrial and fibrillar adhesins and by the expression of heat-stable (ST) and heat-labile (LT) toxins6. ETEC F5 (formerly K99) and F17 fimbriae and ST and LT toxins have been significantly associated with neonatal calf diarrhea (NCD)5. Indeed, ETEC has been considered one of the primary causative agents of NCD5. Enteropathogenic E. coli (EPEC) belongs to the family of pathogens that produce attaching and effacing (AE) lesions when intimately attach to intestinal epithelial cells4. The ability to efface microvilli and to form pedestal-like structures in the intestine is encoded by the pathogenicity island called the locus of enterocyte effacement (LEE)4. Another pathotype that belongs to the AE family is the group known as enterohemorrhagic (EHEC)/Shiga toxin-producing E. coli (STEC). While all the strains belonging to this group are characterized by the expression of Shiga toxin 1 and 2 (individually or simultaneously), some of them produce AE lesions to the intestinal epithelial cells4. EHEC/STEC is a well-documented zoonotic pathogen, responsible for important outbreaks around the world11. Bovines are asymptomatic carriers of STEC, responsible for their spread6. Finally, necrotoxigenic E. coli (NTEC) is characterized by the expression of different virulence factors, including fimbrial and afimbrial adhesins, siderophores, and toxins (cytotoxic necrotizing factor, CNF and cytolethal distending toxin, CDT). So far, NTEC expresses two types of CNF. NTEC1 is frequently isolated from diarrhea in domestic animals and ruminants whereas NTEC2 is mostly associated with septicemia in ruminants6.
Dairy farms are distributed across Uruguay, and the production of milk and dairy products is one of its most important agricultural activities. Pathogenic E. coli associated with NCD in the country has been previously characterized by our group. In that work, several E. coli virulence genes were detected in animals with signs of NCD and in healthy ones, F17 and CS31A adhesin genes being the most prevalent in both groups of calves13.
The aim of this study was to detect the presence of 21 virulence genes, distinctive of the most relevant E. coli pathotypes in bovines: ETEC, EPEC, EHEC/STEC and NTEC, in feces from dairy calves with signs of diarrhea and asymptomatic ones throughout Uruguay. These analyses increased the panel of assessed E. coli virulence genes providing an update of the pathogenic profile of E. coli associated with NCD in dairy farms in Uruguay.
Feces of 252 (149 diarrheic and 103 healthy) calves younger than 35 days-old were processed between 2016 and 2018. Samples were collected throughout the Uruguayan territory by veterinarians and shipped chilled to the laboratory. All samples were plated onto selective MacConkey agar plates (OXOID) within 12h following collection. After 24h of incubation at 37°C, at least 10 lactose positive colonies of each animal were selected and biochemically identified8. Molecular characterization included the evaluation of 21 E. coli virulence genes using conventional and multiplex PCR and previously described primers7,9,15 (supplementary Table). PCR analyses were performed in pools of DNA, at a final concentration of 50ng/μL. Each pool consisted of equal quantities of genomic DNA of E. coli isolates (a maximum of 10 isolates from each animal, see above) of every evaluated animal.
Odds ratio (OR) was performed to measure the association between the presence of the evaluated genes, the occurrence of symptoms, and the geographical origins of the isolates, considering statistical significance when p-values were lower than 0.05.
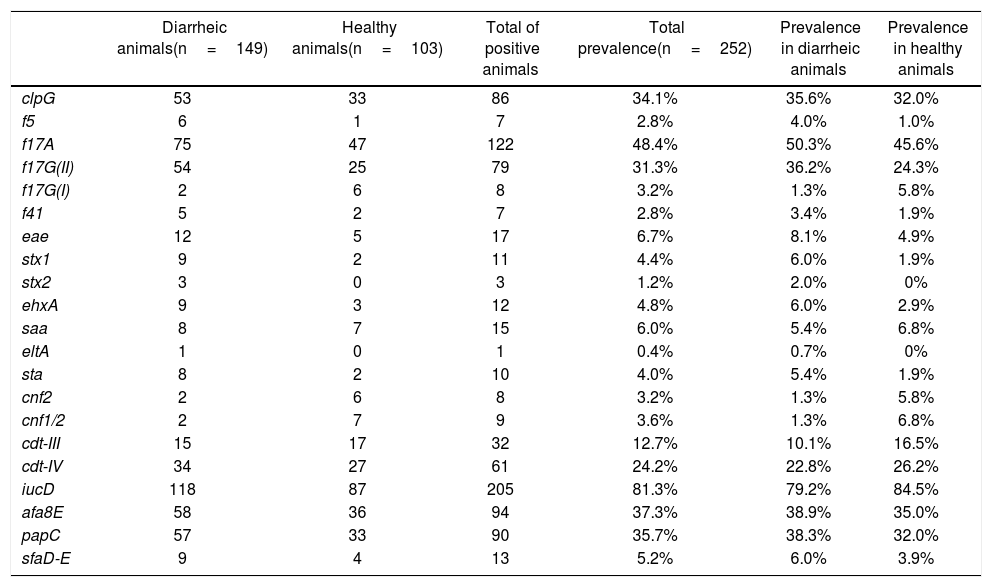
All the evaluated E. coli virulence genes (VGs) were detected in this study. At least 1 animal presented one of the evaluated genes, and some showed more than one gene at the same time (a maximum of 7 VG was detected simultaneously in a pool of DNA). Additionally, all evaluated genes were detected in healthy calves and in calves with signs of NCD, except for Shiga toxin 2 gene (stx2) and heat-labile toxin gene (eltA), which were detected only in diarrheic animals (Table 1). No association between any VGs and animal signs was detected.
Presence of virulence genes in healthy calves and calves with signs of NCD.
| Diarrheic animals(n=149) | Healthy animals(n=103) | Total of positive animals | Total prevalence(n=252) | Prevalence in diarrheic animals | Prevalence in healthy animals | |
|---|---|---|---|---|---|---|
| clpG | 53 | 33 | 86 | 34.1% | 35.6% | 32.0% |
| f5 | 6 | 1 | 7 | 2.8% | 4.0% | 1.0% |
| f17A | 75 | 47 | 122 | 48.4% | 50.3% | 45.6% |
| f17G(II) | 54 | 25 | 79 | 31.3% | 36.2% | 24.3% |
| f17G(I) | 2 | 6 | 8 | 3.2% | 1.3% | 5.8% |
| f41 | 5 | 2 | 7 | 2.8% | 3.4% | 1.9% |
| eae | 12 | 5 | 17 | 6.7% | 8.1% | 4.9% |
| stx1 | 9 | 2 | 11 | 4.4% | 6.0% | 1.9% |
| stx2 | 3 | 0 | 3 | 1.2% | 2.0% | 0% |
| ehxA | 9 | 3 | 12 | 4.8% | 6.0% | 2.9% |
| saa | 8 | 7 | 15 | 6.0% | 5.4% | 6.8% |
| eltA | 1 | 0 | 1 | 0.4% | 0.7% | 0% |
| sta | 8 | 2 | 10 | 4.0% | 5.4% | 1.9% |
| cnf2 | 2 | 6 | 8 | 3.2% | 1.3% | 5.8% |
| cnf1/2 | 2 | 7 | 9 | 3.6% | 1.3% | 6.8% |
| cdt-III | 15 | 17 | 32 | 12.7% | 10.1% | 16.5% |
| cdt-IV | 34 | 27 | 61 | 24.2% | 22.8% | 26.2% |
| iucD | 118 | 87 | 205 | 81.3% | 79.2% | 84.5% |
| afa8E | 58 | 36 | 94 | 37.3% | 38.9% | 35.0% |
| papC | 57 | 33 | 90 | 35.7% | 38.3% | 32.0% |
| sfaD-E | 9 | 4 | 13 | 5.2% | 6.0% | 3.9% |
ETEC genes were detected in low numbers. Genes encoding for heat-stable (sta) and heat-labile (eltA) toxins were present in 10 animals (1 animal presented both toxin genes simultaneously), and only 7 sta+ animals were also f5+. F41 fimbriae were found in a limited number of animals (n=7), where 5 of them were also f5+. Among all adhesins and adhesion-related genes, f17A, afa8E, papC, clpG and f17G(II) were the most abundant (48.4%, 37.3%, 35.7%, 34.1% and 31.3%, respectively). The gene that codifies for the F17A structural subunit was one of the most represented genes in this study, after the iucD gene of the aerobactin operon, whose prevalence was 81.3% (Table 1). On the other hand, the less prevalent adhesion-related genes were saa (STEC autoagglutinating adhesin), sfaD-E (mannose-resistant S fimbriae) and f17G(I) detected only in 15, 13 and 8 animals, respectively (prevalence 6.0%, 5.2% and 3.2%).
Genes associated with the EHEC/STEC group were also poorly represented. Seven of the 252 animals were stx1+/eae+ (6 calves with signs of NCD and 1 healthy animal), 2 animals with NCD signs were stx2+/eae+, 1 animal with signs of NCD was stx1+/stx2+/eae+, 2 animals with NCD signs were stx1+ and 1 animal with signs of NCD was stx2+. E. coli hemolysin gene ehxA, which can be used as an epidemiological marker for EHEC/STEC strains, was identified in 12 isolates (prevalence 4.8%). Seven of these 12 animals were eae+/stx1+/ehxA+, 3 were eae+/ehxA+, 1 was eae+/stx2+/ehxA+ and 1 was eae+/stx1+/stx2+/ehxA+. Likewise, the EPEC distinctive characteristic eae+ was detected in 7 (3 animals with NCD signs and 4 healthy) animals (Table 1).
As mentioned above, iucD was the most prevalent gene in the collection. This aerobactin gene is commonly present in extraintestinal E. coli strains, including NTEC. CNF was present in 11 animals (some animals were cnf2+ and cnf1/2+ simultaneously) and CDT was detected in animals with signs of NCD and in healthy ones, the gene variant cdt-IV being more prevalent than the cdt-III variant (24.2% and 12.7%, respectively) (Table 1).
Approximately 1700 homolog gene clusters form the core genome of E. coli, while the composition of the pangenome of this organism is about 16400 gene clusters3. Variability in genome sizes is the consequence of the remarkable plasticity of the genetic material of this bacterium. The efflux of the flexible gene pool due to transposons, integrons, bacteriophages, plasmids, insertion elements and pathogenicity islands together with mutations, rearrangements, deletions and duplication events result in new combinations of genes and accelerate the emergence of virulent strains. When this combination of VGs persists, the pathotypes emerge4.
F17 (f17A and f17G(II)) fimbriae and CS31A (clpG) afimbrial adhesin, essential in the first steps of attachment to the intestinal epithelium, were the most prevalent encompassing all adhesin and adhesion-related genes evaluated here. Similar results were observed in a smaller collection of E. coli in calf feces in our country in 201613. On both occasions, f17A, f17G(II) and clpG were present in a high prevalence in both groups of animals (with and without signs of NCD). Meta-analyses performed by Kolenda et al.5 statistically demonstrated that F17 is more frequent in diarrheic than in healthy animals; still, a high prevalence of this adhesin in healthy calves is observed. Two assumptions are raised regarding these observations, (i) F17 is not expressed even though the PCR method detects its presence, or (ii) this fimbria requires the presence of other virulence factors to participate in the etiology of NCD5. On the other hand, prevalence of F5 (f5) and F41 (f41), which are highly associated with NCD and are the most investigated ETEC fimbriae, was low (<2% each gene). Similar results were previously reported in Uruguay and regionally12,13. Furthermore, our results are consistent with the observation of several authors who showed a significant decline of f5 over time, mainly attributed to the fact that available vaccines against NCD include this antigen5. Concomitantly with the level of f5 and f41 genes, heat-stable and heat-labile ETEC toxins genes were detected in low numbers.
Serious outbreaks caused by EHEC/STEC strains have been reported around the world, most of the time associated with contaminated raw meat or vegetables11. It is well known that infections caused by these pathogens are serious zoonoses since ruminants, particularly bovines, are the main reservoirs of these strains. In Uruguay, the incidence of hemolytic uremic syndrome (HUS) and hemorrhagic colitis (HC), two important diseases associated with EHEC/STEC infection, is low. It is estimated in 4 out of 100000 children under 5 years old, whereas in Argentina, one of the countries of major incidence of HUS in the world is 12 out of 100000 children1,10. In this work, only 13 animals harbored one of the distinctive genes stx1 and stx2 alone, or in combination with eae. This low prevalence was previously reported by our group and could corroborate the low incidences of EHEC/STEC diseases detected in Uruguay14.
NTEC comprises a pathotype that shares many properties of typical extraintestinal E. coli, including the presence and expression of iron-sequestering systems, various fimbrial and afimbrial adhesins and resistance to the bactericidal action of complement4. Moreover, NTEC is defined by the presence and expression of CNF and CDT toxins and it can be detected in human and animal infections as well as in healthy individuals6. In this work, CNF and CDT toxins genes were detected in animals with or without signs of NCD, cdt-IV and cdt-III being more prevalent than cnf2 and cnf1/2. In addition to the toxins, NTEC strains express factors associated with invasion, which are essential to cause septicemia and internal organ infections6. The iucD gene is a component of the aerobactin operon, and was the most prevalent gene in our collection, mostly detected simultaneously with adhesins (mainly f17A, clpG, afa8E and papC) and NTEC toxin genes. All these results are in accordance with previous works that demonstrate the co-existence of diarrheagenic and extraintestinal E. coli VGs in each calf4,6.
In summary, this study reports updated data about the virulence profile of E. coli in dairy calves in Uruguay, mainly diarrheagenic but also extraintestinal E. coli associated-genes. A large number of adhesins and toxin genes were observed, although no relationships between them and animal symptoms were noted. Therefore, other NCD risks factors such as co-infections, poor nutrition, inadequate colostrum consumption and animal hygiene may probably contribute to the disease outcome. Furthermore, the presence of EHEC/STEC genes exposes the occurrence of potentially zoonotic E. coli isolates in calf feces.
FundingThis work was funded by the project PL-15 from “Instituto Nacional de Investigación Agropecuaria” (INIA), and the project FMV-104922 from the Uruguayan “Agencia Nacional de Investigación e Innovación” (ANII).
Conflict of interestThe authors declare that they have no conflicts of interest.
We are very grateful to all the Farmers who collaborated in sample collection. Authors especially thank Sofía Acquistapace, Cecilia Monesiglio, Verónica Szpinak and Lucía Trujillo for their remarkable collaboration in laboratory work.