Shiga toxin-producing Escherichia coli (STEC) is a group of pathogenic enterobacteria of significant public health importance due to their association with highly prevalent human diseases. STEC is ubiquitous in livestock environments, and its presence in the environment emphasizes the importance of properly managing agricultural effluents to reduce health risks from contamination. In order to detect STEC in the effluent treatment systems of two dairy farms (“A” and “B”) in the southwest of Buenos Aires province, samples (“A”, n=88; “B”, n=72) were taken at two different times of the year (winter and spring) and at various points in the treatment systems. Analysis markers for virulence genes (stx, eae, saa, and ehxA) revealed the presence of STEC in 13.1% of the samples, showing an increase in spring and differences between dairy farms possibly related to their maintenance conditions. After manure, sediments showed the highest proportion of STEC-positive samples, which is relevant due to the ability of these strains to survive in the environment through biofilm formation. Eight genetic profiles were identified among all STEC-positive samples, which are associated with STEC strains that can cause hemolytic uremic syndrome (HUS) and other gastrointestinal diseases. This demonstrates the role of dairy farm environments in the region as reservoirs of pathogenic STEC strains and their impact on public health.
Escherichia coli productora de toxina Shiga (STEC) constituye un grupo de enterobacterias patógenas de gran relevancia en salud pública debido a su asociación con enfermedades humanas de alta incidencia. STEC es ubicua en ambientes pecuarios y su presencia en el medio ambiente destaca la importancia de gestionar adecuadamente los efluentes agropecuarios para reducir riesgos sanitarios por contaminación. Con el objetivo de detectar STEC en los sistemas de tratamiento de efluentes en tambos del sudoeste de la provincia de Buenos Aires, se hicieron muestreos en distintos puntos de ellos en dos explotaciones lecheras (explotación A, n=88; explotación B, n=72) en dos momentos del año, invierno y primavera. El análisis de marcadores de genes de virulencia (stx, eae, saa y ehxA) reveló la presencia de STEC en un 13,1% de las muestras, con un aumento significativo en la primavera y diferencias entre tambos, posiblemente relacionadas con sus condiciones de mantenimiento. La mayor proporción de muestras STEC positivas se encontraron en el estiércol, seguido de los sedimentos, lo que es relevante debido a la capacidad de estas cepas de sobrevivir en el entorno a través de la formación de biofilm. Se identificaron ocho perfiles genéticos entre todas las muestras STEC positivas, los cuales están asociados a cepas de STEC generadoras del síndrome urémico hemolítico (SUH) y de otras enfermedades gastrointestinales. Esto demuestra el papel que desempeñan los establecimientos lecheros de la región como reservorios de cepas patógenas de STEC y su potencial impacto en la salud pública.
Shiga toxin-producing Escherichia coli (STEC) represents a group of pathogenic bacteria associated with foodborne human diseases worldwide. These bacteria are responsible for both sporadic cases and outbreaks of diarrhea, with or without blood, as well as hemolytic uremic syndrome (HUS)23. STEC strains characteristically possess at least one of the Shiga toxin virulence genes, such as stx1, stx2, and their variants. Additionally, they can carry genes for adhesin-intimin (eae), enterohemolysin (ehxA), and/or autoagglutinating adhesin (saa), which enable STEC strains to attach, colonize, and produce toxins26,27,34. There are over 1000 serotypes of STEC identified, among which 400 are associated with human diseases widely distributed around the world4. In Argentina, HUS is endemic and is primarily associated with the O157:H7 serotype (stx1 and/or stx2, eae, ehxA)29. The country reports an annual incidence of 300–400 cases of HUS in children under 5 years old (Ministry of Health, 2017). Additionally, in Argentina, non-O157 STEC strains are responsible for at least 25% of STEC infections7,31.
STEC strains are widespread in farm environments, where healthy cattle and other ruminants naturally carry the pathogen in their gastrointestinal tracts10. Water, sediments, and soil serve as secondary reservoirs for STEC in the environment15,17,30. These pathogens can enter the environment through various pathways, including manure application, runoff from animal farms, and the feces of wild animals1. Once STEC are introduced into the environment, especially in agricultural soils, they can persist for days or even months. This prolonged presence increases the likelihood of contamination in the food chain, thereby posing a significant risk to public health1.
The intensification of dairy farming in Argentina has led to the implementation of sustainable systems for managing large volumes of effluents. As a result, many establishments have incorporated the stabilization pond system as a treatment for these effluents. Furthermore, these effluents can be considered a valuable resource in agricultural production due to their high concentrations of nutrients and organic matter, which help reduce fertilizer costs. However, improper management of these effluents can transform them into significant environmental pollutants16,21.
There is limited data on the persistence and survival of STEC in the agricultural environment, especially on dairy farms at the regional scale. The objective of this study was to demonstrate the presence of STEC in effluent management systems on dairy farms. The results obtained provide new information regarding effluent management systems as reservoirs of STEC in the region and may contribute to the development of strategies aimed at reducing health risks in this type of environment.
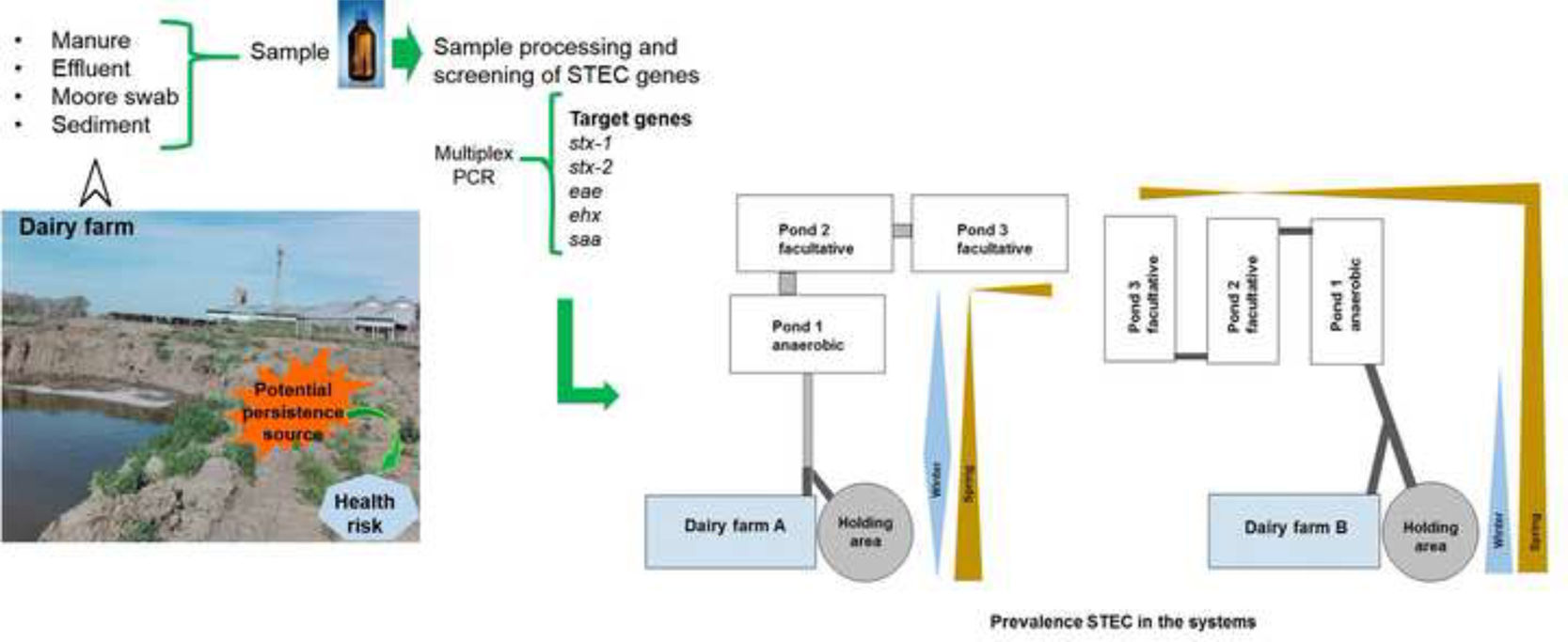
Materials and methodsOrigin and description of the samplesThe two dairy farms, hereafter referred to as “A” and “B”, where the sampling was conducted, are situated in the southwest of Buenos Aires Province, Argentina. Both of these farms employ a stabilization pond system for effluent treatment. Farm ‘A’ maintains a herd of 500 cows, while farm ‘B’ a 1300-cow herd.
The sampling took place from July to December 2017. The average temperature during this period ranged from 8.9°C to 20.8°C. In July and August, there were 15 days of frost, with an absolute minimum temperature range of −5.3°C to −2.2°C. Precipitation levels ranged from 5mm to 69mm during this time frame, with average relative humidity percentages between 58.0% and 82.6% (Table S1).
The effluent treatment system used by the dairy farms consists of three stabilization ponds, with the first pond being anaerobic while the remaining two, though called aerobic, are considered facultative because they have an aerobic surface layer over an anaerobic one. The dimensions of the ponds at each farm are as follows: At “A”, they measure 60m in length, 10m in width, and 2m in depth; and at “B”, they measure 25m in length, 16m in width, and 2–4m in depth.
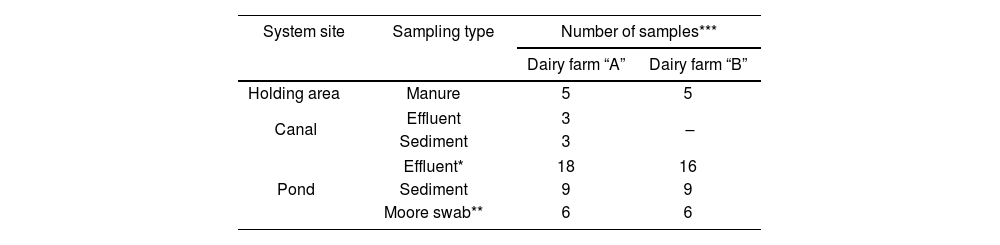
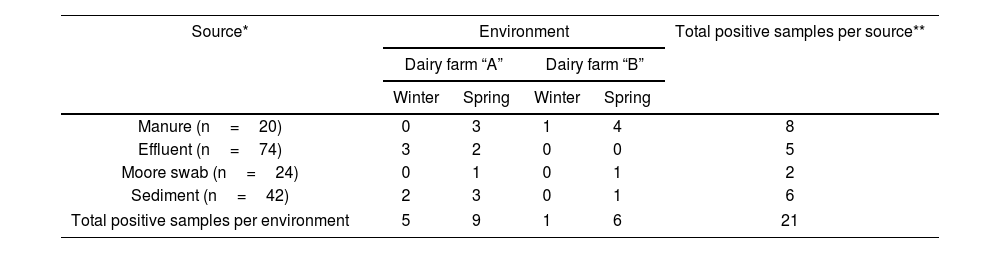
Sampling was conducted in winter and late spring in the year 2017 (August 25th and December 12th). A total of 160 samples of manure, effluents, and sediments were collected. Table 1 provides details on the types of samples collected at each site in both studied systems.
Manure samples were taken from the holding pen using latex gloves, spatulas, and sterile screw-cap PP tubes (50ml). Approximately 500ml of effluents were collected in sterile bottles, and a piped system with a manual water extraction pump was used to access the center of the ponds (Fig. 1). Sediment samples were collected using a dredge and placed in sterile 50ml tubes. Additionally, three Moore swabs were placed in each pond at different depths (Table 1) using a surface buoy anchored 2m from the shore. The swabs were collected after 48h and placed in containers with 250ml of peptone water (Britania, Argentina). All samples were properly refrigerated and processed within 48h of collection.
Sample processing and DNA extractionFive grams or milliliters of each sample were resuspended in 50ml of peptone water (Britania, Argentina) and incubated at 37°C for 24h. Subsequently, the presumptive detection of coliforms was carried out by inoculating 1ml of the enriched culture into a tube containing 9ml of MacConkey broth (Britania, Argentina) with a Durham tube, and incubated at 37°C for 24h. Positive samples (color change to yellow and gas production) were streaked onto Petri dishes containing MacConkey agar (Britania, Argentina) using a loop, and incubated at 37°C for 24h. An aliquot of the confluent growth was inoculated into 8ml of Luria Bertani broth (Britania, Argentina) and incubated at 37°C for 24h. Subsequently, 1ml of the culture was used for DNA extraction using the Quick Bacteria Genomic DNA Extraction Kit (Dongsheng Biotech, China).
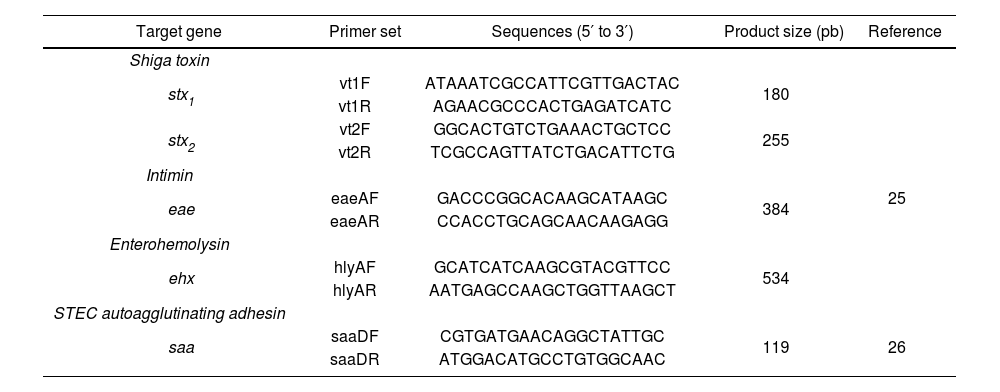
Detection of virulence genes by multiplex PCRA multiplex PCR (mPCR) was performed to screen the samples for the stx, eae, saa, and ehxA genes (Table 2). The O130:H11 strain, provided by the Laboratory of Immunochemistry and Biotechnology of the Faculty of Veterinary Sciences-UNICEN (Tandil, Argentina), carrying the stx1, stx2, ehxA, and saa genes, was used as a positive control. As no specific control strain was available, the presence of eae was inferred from the amplification of a fragment of the expected size (Table 2).
Primers used in PCRm targeting virulence gene.
| Target gene | Primer set | Sequences (5′ to 3′) | Product size (pb) | Reference |
|---|---|---|---|---|
| Shiga toxin | ||||
| stx1 | vt1F | ATAAATCGCCATTCGTTGACTAC | 180 | 25 |
| vt1R | AGAACGCCCACTGAGATCATC | |||
| stx2 | vt2F | GGCACTGTCTGAAACTGCTCC | 255 | |
| vt2R | TCGCCAGTTATCTGACATTCTG | |||
| Intimin | ||||
| eae | eaeAF | GACCCGGCACAAGCATAAGC | 384 | |
| eaeAR | CCACCTGCAGCAACAAGAGG | |||
| Enterohemolysin | ||||
| ehx | hlyAF | GCATCATCAAGCGTACGTTCC | 534 | |
| hlyAR | AATGAGCCAAGCTGGTTAAGCT | |||
| STEC autoagglutinating adhesin | ||||
| saa | saaDF | CGTGATGAACAGGCTATTGC | 119 | 26 |
| saaDR | ATGGACATGCCTGTGGCAAC |
Bacterial DNA amplifications were performed in a reaction volume of 25μl containing 1X Taq Green Buffer; 250nM of primers; 0.2mM (each dNTP) dATP, dGTP, dCTP, and dTTP; 5μl of DNA; and 0.02U of Taq DNA polymerase (Promega, USA). Multiplex PCRs were carried out using a MyCycler thermal cycler (Bio-Rad, USA) with the following program: an initial denaturation at 94°C for 5min, followed by 30 cycles of 94°C for 30s, 60°C for 30s, and 72°C for 45s, with a final extension at 72°C for 5min. The PCR products were separated by electrophoresis on a 2% agarose gel and visualized by staining with GelRed Nucleic acid Gel Stain (Biotium, USA).
Statistical analysisThe data obtained were analyzed using InfoStat software (InfoStat version 2013, InfoStat Group, FCA, National University of Córdoba, Argentina. http://www.infostat.com.ar). Contingency tables and chi-square tests were conducted for STEC-positive samples between the dairy farms (A and B) and between seasons (winter and late spring).
ResultsPresence of STEC in dairy effluent management systemsOut of the total 160 samples analyzed, 21 tested positive (13.1%), indicating the presence of at least one of the virulence genes stx1 and/or stx2. Among these, nine samples (43%) also amplified the ehxA and/or eae genes, while no samples showed amplification of the saa gene.
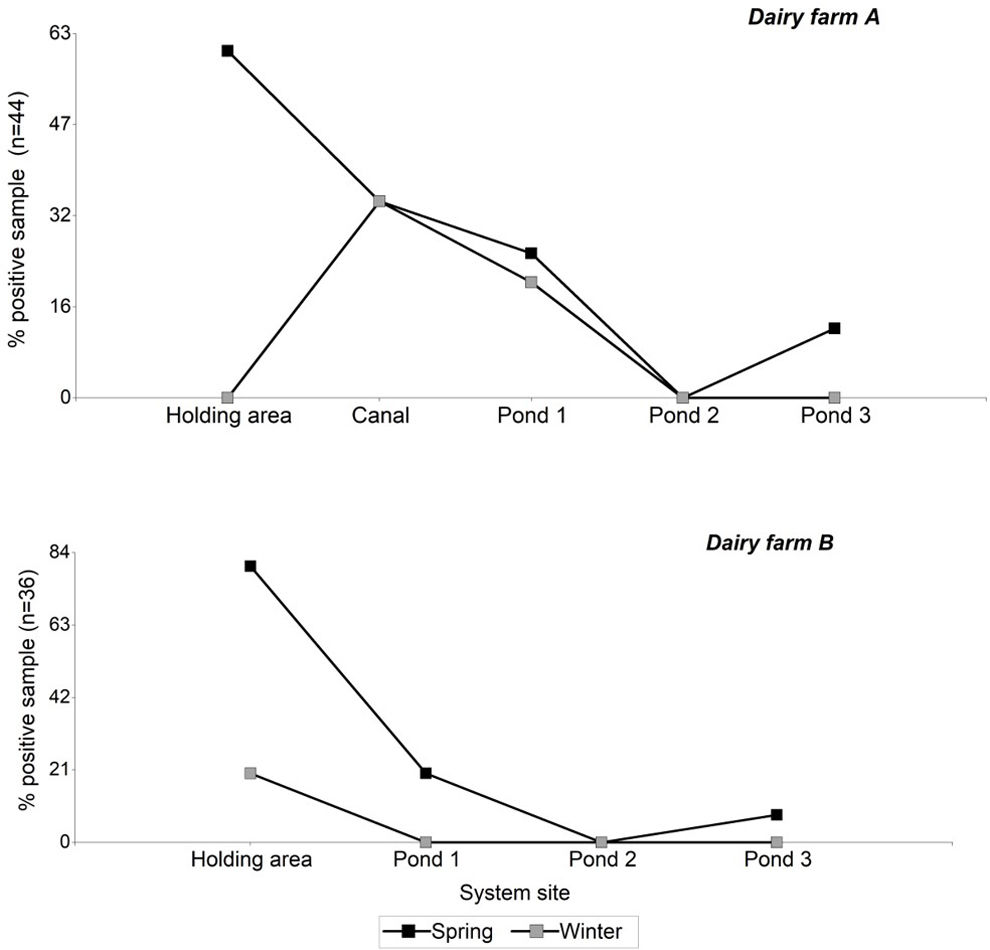
Table 3 describes the number of positive STEC samples according to the source type for each dairy farm and season. Considering the presence of stx genes, the prevalence of STEC was significantly correlated with the season (χ2, p<0.02), with the spring period showing the highest incidence (71% of the total positive samples). While the positive STEC samples did not show a correlation with the dairy farms, dairy farm “A” had a higher number of positive STEC samples compared to dairy farm “B” (66% and 33% of the total positive samples, respectively).
STEC-positive samples according to source type and analyzed environment.
While manure and sediment samples had the highest number of positive STEC samples, no correlations were found with respect to the type of samples analyzed.
Regarding the sites analyzed within each system, it was observed that the presence of STEC decreased from the samples taken from the holding area towards samples collected in the pond 3 (Fig. 2). No stx genes were amplified in pond 2, whereas in pond 3, during the spring season, an increase in positive samples was observed compared to pond 2.
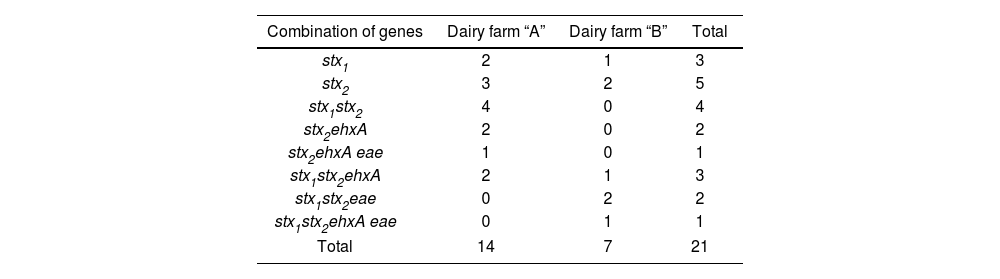
Variability of virulence profilesIn the positive STEC samples, eight combinations of virulence genes were determined (Table 4). The most common genetic profile was the unique presence of the stx2 gene, followed by stx1stx2. Subsequently, different combinations were observed involving the eae and ehxA genes. All the observed combinations included the stx2 gene, making it the most prevalent among STEC genes, found in 18 out of the 21 positive samples. Three profiles, stx1stx2, stx2ehxA, and stx2ehxA eae, were unique to dairy farm “A,” while in dairy farm “B”, two unique profiles were found: stx1stx2eae and stx1stx2ehxA eae.
DiscussionThe intensification of animal production systems has led to the search for sustainable strategies to mitigate the pollution effects resulting from the large quantities of waste generated and to utilize these wastes as resources. In this regard, the stabilization pond system for effluent treatment has been implemented in dairy production establishments to generate a water and fertilization resource while mitigating environmental pollution. However, these types of amendments have been subjected to scrutiny due to their potential as a source of pathogens8,15,19. This has led to the development of tools for implementing good management practices to mitigate contamination.
In the province of Buenos Aires, the presence of STEC strains has been confirmed in cattle from dairy farms at a rate ranging from 13% to 72%12. Additionally, it has been demonstrated that STEC can survive for extended periods in the environment outside the host1,33,38. These data, coupled with the high incidence of HUS cases in the region, underscore the importance of this study. Identifying sources of contamination and reservoirs is crucial for developing management strategies for these establishments, aiming to prevent the spread and contamination of pathogens. Our research group conducted the detection of STEC in effluent treatment systems implemented in two dairy farms in the southwest region of the province of Buenos Aires, with the objective of reusing effluents for fertigation.
It is important to clarify that the detection of stx genes in samples, without the corresponding isolation of host strains, is considered a presumptive determination; however, it is valid for the identification of reservoirs36. In the present study, the detection of stx1 or stx2 genes confirms the presence of STEC (both O157 and non-O157), and the detection of additional virulence genes provides extra epidemiological information about potential pathogenic strains.
By combining growth in culture media and multiplex PCR (mPCR), STEC was successfully detected in various types of samples. The mPCR technique is widely used in the detection and characterization of pathogens and has led to the development of a wide variety of molecular markers for different virulence genes and specific STEC strains3,9,14,25,37. This technique allowed us to identify 13.1% of positive STEC samples, which is consistent with values previously reported for environmental samples30.
While no significant differences were found in the incidence of STEC between the dairy farms, we observed a trend of increased positive STEC samples in dairy farm “A” compared to “B”. This trend could be attributed to infrastructure and management deficiencies observed at the establishments (Orionte S, personal communication). Dairy farm “A” had more precarious facilities and minimal maintenance of the treatment system. Hygiene measures on the farm, as well as effluent management, contribute to the persistence and survival of STEC15,19. Particularly, management can lead to changes in the chemical, physical, and biological characteristics of effluents35.
On the other hand, the seasons of the year show differences in the incidence of STEC. In the spring, there was a higher frequency of positive samples in both establishments for all sites in the analyzed systems. This kind of dynamics has been observed in previous studies for various types of samples (fecal matter, water, sediment, and vegetables)9,12,22. The increase in the incidence of STEC in cattle during the spring compared to winter has been attributed to rising environmental temperatures, rainfall, and other factors such as insect proliferation12. Higher temperatures favor the survival of STEC outside the host12,18. This, in turn, correlates with an increase in cases of HUS24.
In both studied systems, it was observed that the incidence of STEC tends to decrease towards pond 3. Since bovine cattle are the main reservoir of STEC, a higher detection of STEC is expected in manure samples obtained from the holding area. However, on dairy farm “A,” during the winter season, no positive STEC manure samples were detected. The decrease in the presence of stx genes in cattle during the winter season12 may have influenced their detection in this type of samples. Nevertheless, 23% of positive STEC samples were observed in the rest of the system (canal and ponds), indicating the presence of STEC upstream of the system. Later, during the spring season, there was a significant increase in positive STEC samples in the holding area of dairy farm “A,” suggesting a stable presence in the herd.
A particular feature of the effluent management system of dairy farm “A” is the presence of an open 50-meter-long canal that connects the dairy farm and holding area with the first pond. This canal represented the primary reservoir source within the system due to the high incidence of STEC found and its consistency between both seasons. The canal receives effluents from the cleaning of the pen and dairy farm, and experiences some stagnation and the formation of a surface crust layer over much of its course. These conditions may favor the development of a high bacterial load, in addition to the formation of biofilm, facilitating the survival of STEC strains33,40.
There were no positive STEC samples detected in any of the two systems in pond 2; however, the presence of STEC was found in the third pond during the spring season. This dynamic could be explained by external sources of contamination, such as wild animals that increase their activity during spring and summer. The presence of birds, otters, and other rodents has been observed in the area. Wildlife has been identified as vectors of STEC, demonstrating the presence of STEC strains and enteropathogenic E. coli (EPEC) strains related to human diseases2,11,39. However, we would expect this type of contamination to occur randomly for all the ponds. On the other hand, there is evidence of various factors (matrix type, temperature, solar radiation, coexisting microbial communities, effluent management, time, among others) that influence the dynamics of E. coli populations6,13,19,32,35. It is possible that favorable conditions were generated in pond 3, allowing for an increase in bacteria and enabling the detection of STEC. However, the absence of previous studies providing information on the evaluated systems does not allow us to draw conclusions in this regard, creating opportunities for further research.
The incidence of STEC for the different types of analyzed samples (manure, sediment, effluent, and Moore swab) ranged from 5% to 1.2% of the total analyzed, with manure samples showing the highest values, followed by effluent and sediment samples (3.1% and 3.7%, respectively), in line with values reported by other authors9. Of the total sediment samples analyzed, 15% tested positive, which was higher than the positive rate for effluent samples (7%). Cooley et al.9 using various methodologies to detect and isolate STEC in a diverse set of samples from agricultural establishments, found incidences ranging from 3% to 18%, with the highest values observed in sediments. During the spring season, there was an increase in STEC in sediment and manure samples compared to the other sample types. The presence of STEC in pond sediments is noteworthy due to the potential ability of STEC to form biofilms5,20,40, thereby prolonging survival and serving as a latent reservoir of this pathogenic group.
It was interesting to observe that in the establishment with better system maintenance conditions (dairy “B”), no STEC was detected in effluent samples, while under poor conditions and maintenance (dairy “A”), the presence of STEC was observed in the same type of samples during both seasons of the year. Once again, it becomes evident that the management and maintenance of the establishments are key factors that could influence the incidence and persistence of STEC in the environment.
The different virulence factors used for STEC detection allowed for the determination of eight virulence profiles in the analyzed samples. The diversity in these profiles indicates the existence of different pathogenic strains. Although this study did not involve the isolation and serotyping of strains, the amplification of virulence genes predicts the presence of STEC as well as other EPEC strains14,33. Moreover, we must not disregard the presence of other members of the Enterobacteriaceae family that produce Shiga toxin and cause gastrointestinal diseases and HUS in humans, such as Shigella spp., Citrobacter spp., Aeromonas spp., and Enterobacter spp., in which the stx genes have been reported28.
The combination of factors stx2, eae, and ehxA corresponds to the most characteristic profile of the primary strain responsible for HUS, O157:H7. Identifying profiles associated with STEC strains that cause HUS, as well as other enteric diseases, raises concerns about the potential of these establishments as reservoirs of this pathogenic group.
This study is the first to investigate the presence of STEC in the region within a livestock environment and its effluent treatment system. It is essential to gain a better understanding of the prevalent strains in the region and the dynamics of these populations in the evaluated environments as a health tool and for monitoring STEC strains. The low infectious dose of STEC is also important; therefore, even if coliform levels are reduced, a small number of surviving or persistent STEC can still pose a problem23. Characterizing the variability of strains in the area, their correlation with other regions of the country, and their persistence in these environments and agricultural soils are questions that need to be addressed.
In general, pond-based effluent treatment systems are efficient and have been shown to reduce the most probable number of coliforms21, allowing for their utilization as water and fertilization resources under specific management conditions32. However, in Argentina, there is still a need for studies regarding the presence and prevalence of STEC in amendments, as well as the evaluation post-application of these organic materials in agricultural soil.
In conclusion, the study highlighted the persistence of STEC strains in the effluent treatment systems of two dairy farms in the southwest of the Buenos Aires province. The data obtained reaffirm the role that dairy environments in the region play as reservoirs of pathogenic STEC strains and underscore a problematic focus for the healthcare system. Given the high incidence of STEC-associated diseases in this region, it is expected that the provided information will be relevant to public health, offering a tool for the development of intervention strategies on dairy farms aimed at reducing both direct and indirect human exposure to STEC, lowering the burden in the cattle herd, and enabling the reuse of effluents as an agricultural resource, thus creating a sustainable system.
Conflict of interestThe authors declare that they have no conflicts of interest.
We express our appreciation to the farm owners and farm personnel for their collaboration in this study; to Ing. Agr. Sebastían Orionte (INTA Hilario Ascasubi) for her connection with farms. To Drs. Nora Padola and Daniel Fernández (Laboratorio de Inmunoquímica y Biotecnología, Facultad de Ciencias Veterinarias-UNCEN) for valuable collaboration in facilitating laboratory protocols and providing strain O130:H11. This investigation was supported by the Agencia Nacional de Promoción Científica (ANPCyT, PICT 2014-1760 and PICT 2020-02289) and the Secretaría de Ciencia y Técnica – UNS (PGI 24/A250).