Rectal swabs (122) from pediatric patients were analyzed by polymerase chain reaction (PCR) for the detection of EPEC and STEC. STEC isolates were tested for the presence of stx1, stx2, eae, saa and ehxA. All eae-positive samples were tested for the presence of bfpA, and antigen O was determined using the agglutination test. Int1 and Int2 were detected to identify the presence of integrons class 1 and 2, respectively. Escherichia coli was detected in 68% of the samples, of which 18.8% were STEC (2.45%) and EPEC (16.3%). Serogroups STEC O145 and EPEC O130, O113 and O157 were observed, while three strains were non-typable. None of the EPEC strains carrying tbfpA and class 1 and 2 integrons was detected in any of the samples. The results obtained are important considering the virulence profiles found in the isolated EPEC and STEC strains and the serogroups associated with disease in humans.
Se analizaron hisopados rectales (n=122) de pacientes pediátricos mediante reacción en cadena de la polimerasa (PCR) para detectar cepas de E. coli enteropatógenas (EPEC) y productoras de toxina Shiga (STEC). En los aislamientos de STEC, se investigó la presencia de los genes stx1, stx2, eae, saa y ehxA. En todas las muestras positivas para eae se evaluó la presencia de bfpA y el antígeno O se investigó mediante aglutinación. Además, se analizó la presencia de Int1 e Int2 para identificar integrones de clase 1 y 2, respectivamente. Se detectó E. coli en el 68% de las muestras, de las cuales el 18,8% fueron STEC (2,45%) o EPEC (16,3%). Se detectaron los serogrupos STEC O145 y EPEC O130, O113 y O157, mientras que tres cepas no fueron tipables. No se detectó ninguna cepa de EPEC que portara tbfpA o integrones de clase 1 y 2 en ninguna de las muestras. Los resultados son importantes considerando los perfiles de virulencia encontrados en cepas aisladas de EPEC y STEC y los serogrupos asociados con enfermedades en humanos.
Diarrheal infectious diseases are the leading worldwide cause of infant morbidity and mortality in children under 5 years of age. Viruses, bacteria, and parasites are the most commonly detected etiological agents. While among viral agents, rotavirus is the leading cause of acute diarrhea in children, the role of bacteria in causing childhood diarrhea appears to differ by geographic area11. Escherichia coli is a bacterium frequently associated with diarrhea, which can be classified into different pathotypes based on their virulence factors and the disease it causes. Enteropathogenic E. coli (EPEC) and Shiga toxin-producing E. coli (STEC) are two pathotypes associated with intestinal diseases that are known as diarrheagenic E. coli (DEC)12.
EPEC can range from subclinical to fatal infections, and is classified into typical EPEC (tEPEC) and atypical EPEC (aEPEC), based on the presence or absence of the virulence-associated EAF plasmid (pEAF), respectively2,9. The pEAF encodes the bundle-forming pilus (BFP) that mediates EPEC autoaggregation and microcolony formation on the surface of epithelial cells, a phenotype known as localized adherence2. Both groups of EPEC produce attaching and effacement (A/E) lesions, derived from the cooperative action of proteins encoded in an island of pathogenicity called locus of enterocyte effacement (LEE)2,3,9. tEPEC strains cause profuse secretory diarrhea with mucus and significant losses of water and electrolytes in the stool, in addition to severe nutrient malabsorption. EPEC transmission occurs by fecal–oral route through contaminated hands, water, and food2. Although tEPEC was described in humans, Sanches et al. discovered the presence of this bacterium in Psittaciformes birds15, and aEPEC was described in animals and humans reservoirs, both also differing in genetics, serotypes, and virulence properties8. aEPEC is most closely related to STEC and appears to be an emerging pathogen8.
STEC can produce bloody diarrhea, hemorrhagic colitis (HC) and hemolytic uremic syndrome (HUS) in humans, and most outbreaks have been attributed to O157:H7 rather than to non-O157 serotypes1,13. In Argentina, HUS is endemic with a median of 314 new cases per year reported in the period 2014–2018, with an annual incidence of 6.52 cases per 100000 children under 5 years of age. HUS is one of the most important causes of acute kidney injury in children and the second most frequent cause of chronic kidney disease in our country1. Cattle are the main reservoir of STEC and the transmission to humans occurs through the consumption of undercooked meat, non-pasteurized dairy products, and vegetables or water contaminated with feces, direct contact with animals or the environment14,15. STEC are classified by the presence of virulence genes, such as those encoding Shiga toxins (stx1, stx2), intimin (encoded by eae), and enterohemolysin (ehxA), although LEE negative strains can carry the autoagglutinating adhesin (saa)14,16.
The increasing level of antimicrobial-resistant bacterial infections in both humans and animals is continuing to cause concern among veterinarians and medics. Class 1 and 2 integrons are important resistance mechanisms and genetic cassettes that contain mobile genetic elements5.
Considering the important role of EPEC and STEC as causes of diarrhea in children, the aim of this study was to detect EPEC and STEC in stools samples from child patients with diarrhea and investigate the presence of virulence genes stx1, stx2 and eae associated with disease in humans as well as to determine the presence of Int1 and 2, as a mechanism of antimicrobial resistance.
From January to March 2019, and February 2020, stool samples from 122 children diagnosed with acute diarrhea at Hospital de Niños Dr. D. Blanco Villegas, from Tandil city and Centro de Medicina Infantil A. Lacroze de Fortabat, from Olavarría city, Buenos Aires, Argentina, were collected. The samples were taken using sterile swabs following standard procedures for routine diagnostics, transported in Cary-Blair transport medium, and immediately processed in the laboratory.
Swabs were cultured in Luria Bertani (LB) broth with shaking at 37°C for 18h. An aliquot (5μl) was diluted in 500μl sterile double-distilled water and boiled for bacterial DNA extraction.
The samples were screened by multiplex PCR to detect stx1, stx2, and eae as indicators for STEC or EPEC presence. E. coli O157:H7 strain EDL933 (kindly supplied by Dr. J. Blanco, Reference Laboratory for Escherichia coli, Lugo, Spain) was used as positive control and double-distilled water, as negative control. Samples eae-positive but stx-negative are considered EPEC, and samples stx-positive but eae negative are considered STEC. Those presenting both stx and eae could be STEC and EPEC or STEC harboring eae.
Samples positive for stx and eae were frozen at −70°C for further isolation of individual stx/eae-positive colonies.
From each stx-positive and eae-positive sample, about 10–200 colonies were analyzed by PCR to detect usp, stx1 and stx2 or eae, for isolation of STEC and EPEC. Each STEC isolate was also tested for the presence of stx1, stx2, eae, saa, and, ehxA by multiplex PCR, according to the experimental conditions described by Rivero et al.14 and usp for identification of E. coli10.
All eae-positive samples were tested for the presence of bfpA by monoplex PCR2. E. coli O157:H45 (bfpAþ) (kindly supplied by Dr. A. Bentancor, Facultad de Ciencias Veterinarias, Universidad de Buenos Aires, Argentina) was used as positive control, and double-distilled water as negative control. Int1 and Int2 were detected. The identification of the somatic antigen O was performed using agglutination with specific antisera provided by the National Institute for the Production of Biologicals-ANLIS Dr. Carlos G. Malbrán.
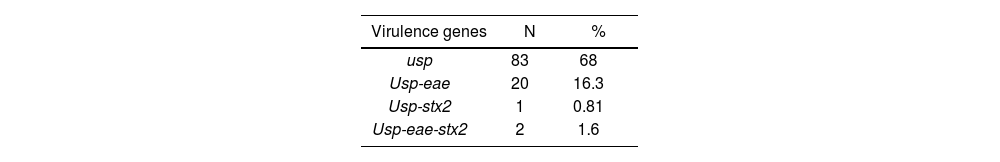
One hundred and twenty-two samples from pediatric patients (80.5% were children under 5 years old, 49% belonged to male patients and 51% to female patients) were analyzed by PCR for the detection of virulence genes from EPEC and STEC. E. coli (usp+) were detected in 68% of the stool samples of patients with diarrhea, of which 18.8% (n=23/122) were STEC/EPEC (16.3% belonged to EPEC and 2.45% to STEC) (Table 1).
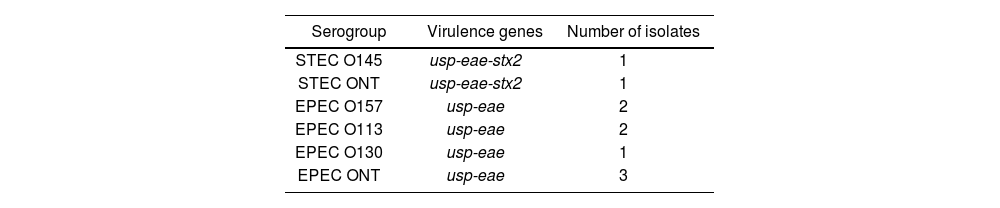
Of the total of positive samples, 43.5% (n=10/23) were characterized, 8 corresponding to the EPEC and 2 to the STEC. The serogroups STEC O145 (1) and EPEC O130 (1), O113 (2) and O157 (2) were detected, while three strains were non-typable (Table 2). None of the EPEC strains carried bfpA. Class 1 and 2 integrons were not detected in any sample.
The frequencies of infection by DEC vary widely according to the geographic region, with values that range from 1% to 52%9. In the present study, 18.8% of the samples were positive for STEC and EPEC, a higher percentage than that obtained by Molina et al. (12.3%) in patients with diarrhea less than 15 years of age. The identified pathotypes were EAEC, ETEC, EPEC and STEC (stx2 and eae)9. Esquivel et al. found that 31% of the stool samples from children were positive for DEC7, and Quiroga et al. obtained 33% of positive samples in children under 20 months12. This difference between studies may be due to the diversity of methodologies, techniques, patients’ age, time of year in which the study was conducted, geographic region, duration of diarrhea, among others. EPEC was detected in 16.3% of the samples, in agreement with other studies from Argentina7,12. All strains were aEPEC, however, in other countries, childhood diarrhea is caused by tEPEC serotypes; however, in developing countries, recent studies have not identified a significant association between tEPEC and childhood diarrhea3,11. Nonetheless, other studies still report that tEPEC is more common than aEPEC as a cause of diarrhea3. Several studies have shown that patients with aEPEC present with mild, non-dehydrating, non-inflammatory diarrhea, being the duration of diarrhea in patients infected with aEPEC generally significantly longer than that caused by other pathogens3. These findings indicate that aEPEC may have an innate propensity to persist longer in the intestine than other diarrheagenic E. coli which cause diarrhea that is more transient in nature. EPEC adheres tightly to epithelial cells and disrupts normal cellular processes12. Moreover, some evidence suggests that aEPEC may decrease apoptosis of intestinal epithelial cells possibly because of the lack of BFP9. A wide variety of EPEC serotypes have been described worldwide2. One of the EPEC serogroups detected in this study was O157. Serogroup O157 contains, in addition to serotype STEC O157:H7, heterogeneous strains, generally not H7, some of which were characterized as EPEC14. In the present work, serogroup O113 was detected in two strains. In Brazil, dos Santos et al., detected two distinct pathotypes of DAE, EAEC and STEC, in association with O113 strains isolated from human and non-human sources, respectively6.
In this study, 2.45% of the samples were positive for STEC. Rivero et al. and López et al., in Argentina, reported a frequency of STEC infections of 10.06% and 4.1%, respectively8,14. Rivas et al. reported a frequency of STEC of 4.1%, with the highest frequencies found in the Southern Region of Argentina (Neuquén 8.7% and Bahía Blanca 7.9%)13. Molina et al., detected a frequency of STEC close to 0.5% in a pediatric outpatient population with diarrhea in La Plata, Argentina9. Other studies conducted in several countries around the world have reported values between 0.3 and 13%9. The variation of the detection range of STEC in cases of diarrhea in children may be due to inclusion criteria, diagnostic method, or socioeconomic factors, among others. In the present study, the isolated STEC were stx2 and eae-positive, which is consistent with the work carried out in Argentina by other authors1,9, and one STEC strain only harbored stx2. Although eae has been reported to play an important role in STEC adherence to enterocytes, different STEC strains lacking eae have been isolated from outbreaks and sporadic cases of diarrhea and HUS16. For this reason, it is currently considered that the presence of the eae gene would not be essential for pathogenesis. The STEC serogroup detected in this study was O145. In addition to the more studied O157:H7 serotype, other non-O157 STEC serotypes, such as O26, O111, O145, O103, and O121, have also emerged with an important role in human disease4. STEC belonging to the O145 serogroup, including O145:NM, O145:H28, O145:H25, O145:H34, O145:H8, O145:H16, and O145:HNT, have been associated with acute watery diarrhea, bloody diarrhea, HC and HUS4. In Argentina, STEC O145 strains are of utmost importance, being the second serogroup after O1574. Despite the importance of DEC, they are not routinely tested in clinical laboratories and proper surveillance is necessary to control and prevent transmission of EPEC and STEC to humans. Advances in the knowledge of pathogenesis, virulence and risk factors have contributed to the development of several strategies aiming to prevent transmission to humans. The results of this work are important considering the virulence genes and the serogroups associated with disease in humans. Therefore, it is important to conduct characterization studies of the isolates, analyzing other genes that may intervene in the pathogenesis. Additionally, it is crucial to consider different variables that could affect the level of susceptibility to infection by EPEC and STEC in children.
Ethical approval and consent to participateAll samples were collected with the legal permission of the parents and information for each patient, such as age, sex, and clinical symptoms, was gathered. Data obtained in this study were analyzed anonymously, ensuring patient confidentiality; ethical approval was not required, and the conditions of this study were reviewed in accordance with the precepts established in the Declaration of Helsinki. In addition, this work was carried out according to the regulations laid down in the Human Health Research Guideline (Resolution 1480/11, Ministry of Health, Argentina).
Conflict of interestThe authors declare that they have no conflicts of interest.