Members of the Pleosporaceae family are known as important sources of airborne allergens which are responsible for asthma and allergic diseases.
AimsThe purpose of this study was to investigate the gene profiling and expression pattern of Alt a 1 in Alternaria alternata and other members of the Pleosporaceae family including Stemphylium botryosum, Ulocladium chartarum, Curvularia lunata, Cladosporium cladosporioides, and Epicoccum nigrum.
MethodsAlternaria alternata and related genera were cultured on Czapek–Dox broth medium at 25°C for 21 days. The presence of Alt a 1 was assessed in fungal culture filtrates by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and then confirmed by immunoblot analysis. Real-time PCR was carried out for quantitation of the Alt a 1 gene encoding corresponding protein at the transcriptional level using cDNA prepared from fungal RNA.
ResultsSDS–PAGE showed protein bands ranging from 14 to 100kDa. A 14kDa band corresponding to Alt a 1 was present in A. alternata, S. botryosum and U. chartarum. The gene expression of Alt a 1 was reported in A. alternata and some other related genera. The Ct mean value recorded for A. alternata strains ranged from 24.70 to 27.84 while it was in the range 23.62–32.09 for other related taxa. No apparent transcription or expression was revealed in C. cladosporioides.
ConclusionsThe presence and efficient expression of Alt a 1 gene in A. alternata and other related taxa indicate that Alt a 1 protein is a major component of the secretory machinery of Pleosporaceae family members, and it may play a crucial role in its allergenicity.
Los miembros de la familia Pleosporaceae son una fuente importante de alérgenos aéreos causantes de asma y enfermedades alérgicas.
Objetivos El objetivo de este trabajo fue estudiar el perfil de expresión génica de la proteína Alt a 1 en Alternaria alternata y otros miembros de la familia Pleosporaceae, entre las cuales pueden citarse Stemphylium botryosum, Ulocladium chartarum, Curvularia lunata, Cladosporium cladosporioides y Epicoccum nigrum.
MétodosAlternaria alternata y otros géneros relacionados se cultivaron en caldo Czapek-Dox a 25°C durante 21 días. La existencia de Alt a 1 en los filtrados de los cultivos se evaluó mediante electroforesis en gel de poliacrilamida con dodecilsulfato sódico (SDS-PAGE) para después confirmarla mediante inmunotransferencia. Se realizó RCP en tiempo real para la cuantificación de la transcripción del gen responsable (Alt a 1) utilizando ADNc a partir del ARN del hongo.
ResultadosMediante SDS-PAGE se visualizaron bandas de proteínas de 14 a 100kDa. La banda de 14kDa, correspondiente a Alt a 1, estaba presente en A. alternata, S. botryosum y U. chartarum. Se detectó expresión génica de Alt a 1 en A. alternata y otros géneros relacionados. El valor medio de Ct registrado en los aislamientos de A. alternata varió entre 24,70 y 27,84; en otros taxones cercanos, el intervalo estuvo entre 23,62 y 32,09. No se detectó transcripción o expresión en C. cladosporioides.
ConclusionesLa presencia del gen Alt a 1 y su expresión en A. alternata y otros taxones próximos indica que la proteína Alt a 1 es uno de los componentes principales del mecanismo secretorio de los miembros de la familia Pleosporaceae y puede desempeñar un papel fundamental en su capacidad alergénica.
Fungal spores are an important part of airborne microflora and are widely distributed in air, soil and other natural environments.20,25,32,36 Fungal allergy is a global problem affecting 4–41% of the human population depending on the country, city, region, age, sex and other factors.7,26 It has been demonstrated that fungi belonging to the genera Alternaria, Aspergillus, Penicillium, Cladosporium, Curvularia, Epicoccum, and Stachybotrys play an important role in the etiology of fungal allergy worldwide.20,32,33,35 Nonetheless, about 150 individual fungal allergens have been identified from approximately 80 mold genera.25,36 The genus Alternaria is considered to be one of the most prolific producers of fungal allergens. It has been shown that Alternaria alternata is a major aeroallergen in many parts of the world, even contributing to asthma attack.8,16,21,25,38 At present, a total of 17 allergenic proteins are characterized as allergens of A. alternata and listed in allergen platforms (http://www.allergen.org/andhttp://www.allergome.org/). In particular, Alt a 1, the major allergen produced by Alternaria species, has been associated with asthma.4,13,15,40 Alt a 1 is a 30kDa-dimer glycoprotein with a unique cysteine-linked β-barrel fold, and composed of two subunits of 16.4 and 15.3kDa; the protein is located in the cytoplasm of fungus spores and mycelia, and its biological function in cells is still mysterious.9–11 Alt a 1 is an excellent marker for measuring sensitization to Alternaria in allergic patients and can be used for therapeutic purposes.4,34,40 Despite the established role of this protein in fungal allergies, more studies are necessary to evaluate the environmental exposure to this allergenic protein because low (0.4%) and high (87%) frequencies of Alt a 1 detection have been reported in dust and air.31,39 Alt a 1 is a conserved protein that is highly specific for Alternaria, while homologs of Alt a 1 have been reported in other Pleosporaceae species.1,18,27 When there is an allergy to A. alternata a very significant level of allergen cross-reactivity to other fungi belonging to the Pleosporaceae family, including Stemphylium, Ulocladium, and Curvularia, can be present, which may be related in part to the expression of different levels of Alt a 1 in these genera.17,30 The analysis of the conservation of the nucleotides in Alt a 1 homologs might be valuable to understand the evolution of this gene and its potential role in fungal biology. 18,37
The purpose of this study was to evaluate the presence and expression profile of Alt a 1 in members of the Pleosporaceae family other than A. alternata, and to compare the expression level of the Alt a 1 encoding gene in various genera and species which are shown to be well correlated with the fungal allergenicity.
Material and MethodsFungal strains and culture conditionsAlternaria alternata PTCC 5224, Stemphylium botryosum FMR 3952, Ulocladium chartarum ATCC 18044, Curvularia lunata CBS411085, Cladosporium cladosporioides FMR 5318, Epicoccum nigrum CECT 2848, and airborne isolates of A. alternata (TA24, TA27, TA29 and TA35) were obtained from the Pathogenic Fungi Culture Collection of the Pasteur Institute of Iran and Laboratory of Mycology, Department of Medical Sciences, Rovira i Virgili University, Tarragona, Spain. Penicillium chrysogenum PTCC 5034 and Aspergillus fumigatus AF-54 were used as controls. All fungal strains were cultured on potato dextrose agar (E. Merck, Germany) and incubated at 28°C for 7 days. Fungal spore suspension was prepared in 0.1% Tween 80 solution and counted on a Neubauer slide. All fungi were cultured on Czapek–Dox broth (2×106 spores/mL) and incubated at 25°C for 21 days in the stationary condition.
Alt a 1 assay in fungal culture filtratesThe presence of Alt a 1 in fungal culture filtrates was assessed as described by Martinez et al.23 Briefly, culture filtrates (LCF) from Czapek–Dox broth medium were filtered through Whatman filter paper No. 1 (Whatman Int. Ltd, Maidstone, England) and sterilized with a glass-fiber prefilter (Millipore, Bedford, MA, USA). The retained material was dialyzed by ultrafiltration (Millipore, USA) with a 10,000Da cut-off point filter paper and concentrated by freeze-drying technique. The protein content of LCF was measured spectrophotometrically at 595nm according to the Bradford method.5
Determination of Alt a 1 by gel electrophoresis and western blottingSDS–PAGE was performed according to the method of Laemmli22 on a 12.5% v/v resolving gel (3M Tris–HCl, pH 8.8) and 5% v/v stacking gel (0.5M Tris–HCl, pH 6.8) in a discontinuous buffer system (Tris 0.025M, Glycine 0.192M, SDS 0.1% (w/v), pH=8.3). Desirable amounts of LCF from all the fungi (8mg/mL), and purified rAlt a 1 (Biomay, Vienna, Austria) were loaded on SDS–PAGE under the abovementioned conditions. The separated proteins were transferred electrophoretically to polyvinylidene fluoride (PVDF) membranes (Immobilon-P, Millipore) according to Asturias et al.3 The membranes were rinsed in Tris buffered saline with Tween® 20 (TBST; 10mM Tris HCl [pH=8.0], 150mM NaCl, and 0.05% Tween 20), with 1% dried milk and incubated with rabbit anti-rAlt a 1 IgG (1:5000 dilution, Bial Laboratorios, Spain) at 4°C overnight. The membranes were washed and incubated with horseradish peroxidase-conjugated swine anti-rabbit immunoglobulin G antibody (Dako, Glostrup, Denmark) (1:10,000 dilution in TBST) to detect the specific band of Alt a 1. The membranes were washed 3 times and developed with the enhanced chemiluminescence (ECL) western blotting detection system (western blotting detection reagents, Amersham Biosciences). The PVDF membranes were rinsed in orthophenylenediamine (OPD; Sigma-Aldrich, USA). The reaction was stopped by adding H2O2 and was exposed to X-ray film for 10s to 10min depending on the intensity of the signal.3
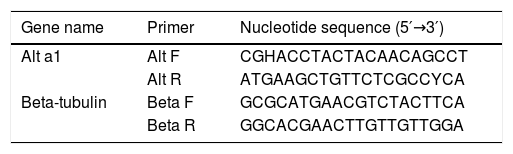
cDNA synthesis and sequencingFungal strains (2×106 spores/mL) were cultured on Czapek–Dox broth medium at 25°C for 21 days. Fungal RNA was extracted from mycelia according to Jahanshiri et al.19 Briefly, 100mg of mycelia were separated by cheesecloth and ground using a sterile mortar and pestle. Total RNA was extracted with the RNeasy Plant Mini Kit (Qiagen, Germany), including a step of genomic DNA digestion with RNase-free DNase, according to the manufacturer's instructions. Concentration and purity of fungal RNAs were determined using a Nano Drop 1000 spectrophotometer (Thermo scientific), and the integrity of each RNA sample was examined with a 1% agarose gel. RNAs were stored at −80°C before use. Complementary DNA (cDNA) was synthesized from 2μg total RNA as the template using M-MuLV reverse transcriptase (Fermentase, Germany) and random hexadeoxynucleotides as primers. Complementary DNA targets were added to a reaction mixture consisting of 10μL, 2×master mix and 0.8μL Alt a 1 primers (5′-GGAACCTACTACAACAGCC-3́; 5′-GTACCACTTGTGGTCCTCAA-3́) designed by AlleleID v.7 (Table 1). The reaction consisted of three stages including stage 1 (initial denaturation), 94°C for 10min; stage 2, 94°C for 30s, 60°C for 30s, and 72°C for 30s; stage 3, 72°C for 5min. Stage 2 was repeated for 35 cycles, and the reaction set also included RT negative control. Agarose gel electrophoresis (1.8%) was performed to confirm the correct size of amplicons with 129bp size and the absence of non-specific bands. Sequencing reactions were performed by using BigDye terminator technology (ABI, Foster City, CA) with an ABI Prism 3730 (ABI series) DNA sequencer. Alt a 1 locus was sequenced in both forward and reverse directions with the same primers mentioned above. Nucleotide sequences were defined by the alignment of forward and reverse sequences using MEGA software version 6.0 and compared with the GenBank database. Polymorphic sites were confirmed by visual examination of the chromatograms.
Real-time PCR analysis of Alt a 1-encoding geneTo calibrate the qRT–PCR system, standard curves from serial dilutions (10−1 to 10−1)[Query:……] of the cDNA template from A. alternata were used. These also served as positive controls for primer and encompassed the entire concentration range of the samples measured. Real-time PCR was carried out using the SYBR green master mix (Applied Biosystems), in a final volume of 25μL for each reaction, by ABI PRISM 7500 thermal cycler (Applied Biosystems). Every experiment was repeated three times. The relative expression ratio (RQ) of Alt a 1 (target) was calculated in comparison to a reference gene (β-tubulin as endogenous control). ΔCT was calculated using the following formula: [ΔCT=CT (target) − CT (reference)] and was performed using the comparative threshold cycle method (2−ΔΔCT) between samples and calibrator (A. alternata PTCC 5224).19
Statistical analysisThe data of gene expression were analyzed by One-way ANOVA using GraphPad Prism 6 (GraphPad Prism Software Inc, SanDiego, CA, USA). P-values lower than 0.0001 were considered significant.
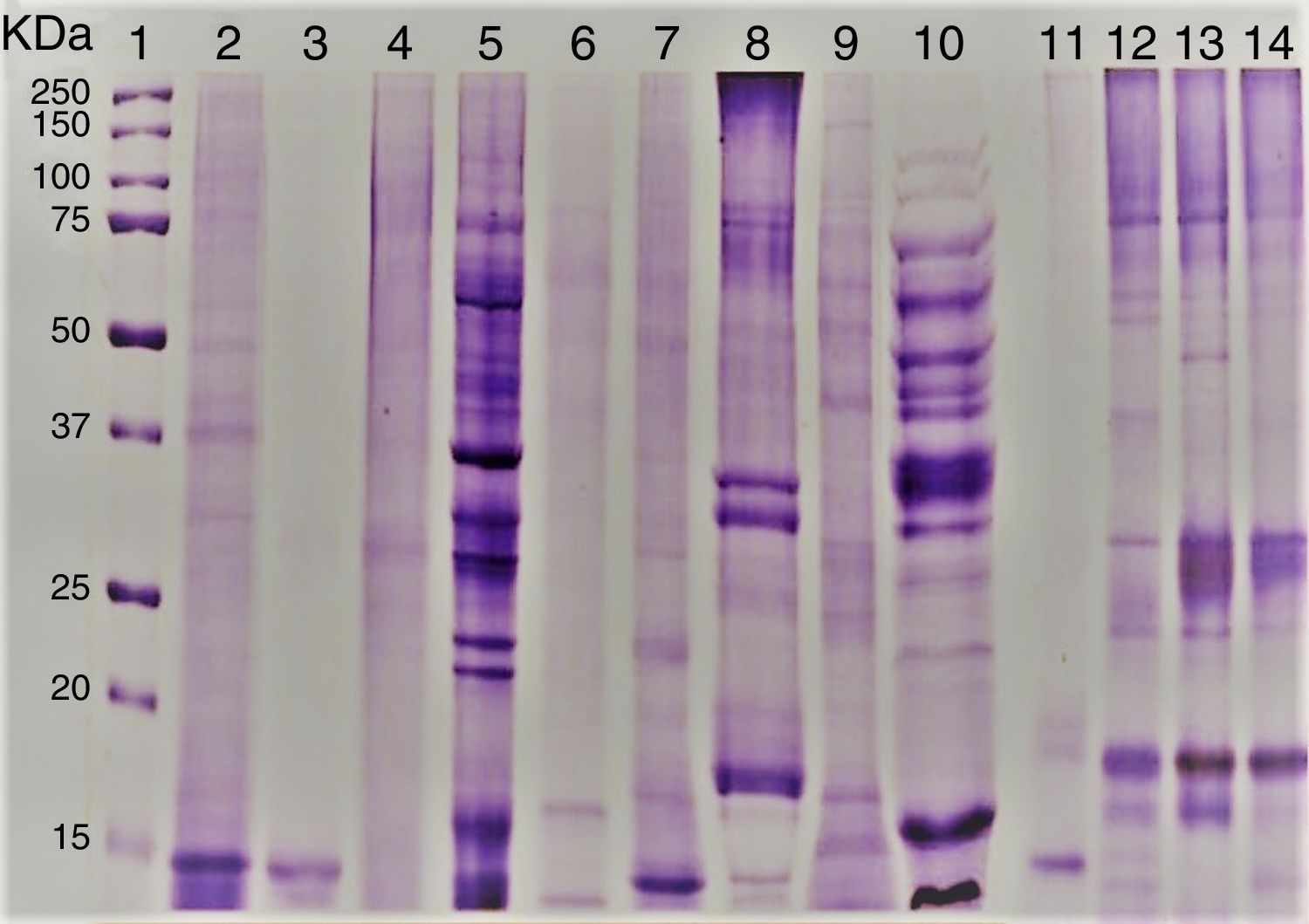
ResultsEvaluation of Alt a 1 protein by gel electrophoresis and western blottingThe amount of proteins present in the fungal culture filtrates from airborne A. alternata and other standard species was in the range 0.5–40mg/L. SDS–PAGE analysis showed that Alt a 1 was separated into 15 and 16kDa subunits under reducing conditions. rAlt a 1 migrated as an approximately 15kDa protein under reducing conditions but tends to dimerize during storage and shows both 29 and 15kDa bands under non-reducing conditions (Fig. 1). The electrophoretic banding patterns of pleosporales (A. alternata, S. botryosum, U. chartarum, and C. lunata) and dothideales (C. cladosporioides) revealed a variable number of bands, ranging from 14 to 100kDa (Fig. 1).
SDS–PAGE of A. alternata and related genera; molecular weight marker protein (1), A. alternata PTCC 5224 (2), rAlt a 1 protein (3), E. nigru (4), U. chartarum (5), S. botryosum (6), C. lunata (7), C. cladosporioides (8), A. fumigatus AF-54(9), P. chrysogenum (10) and A. alternata airborne strains (11–14).
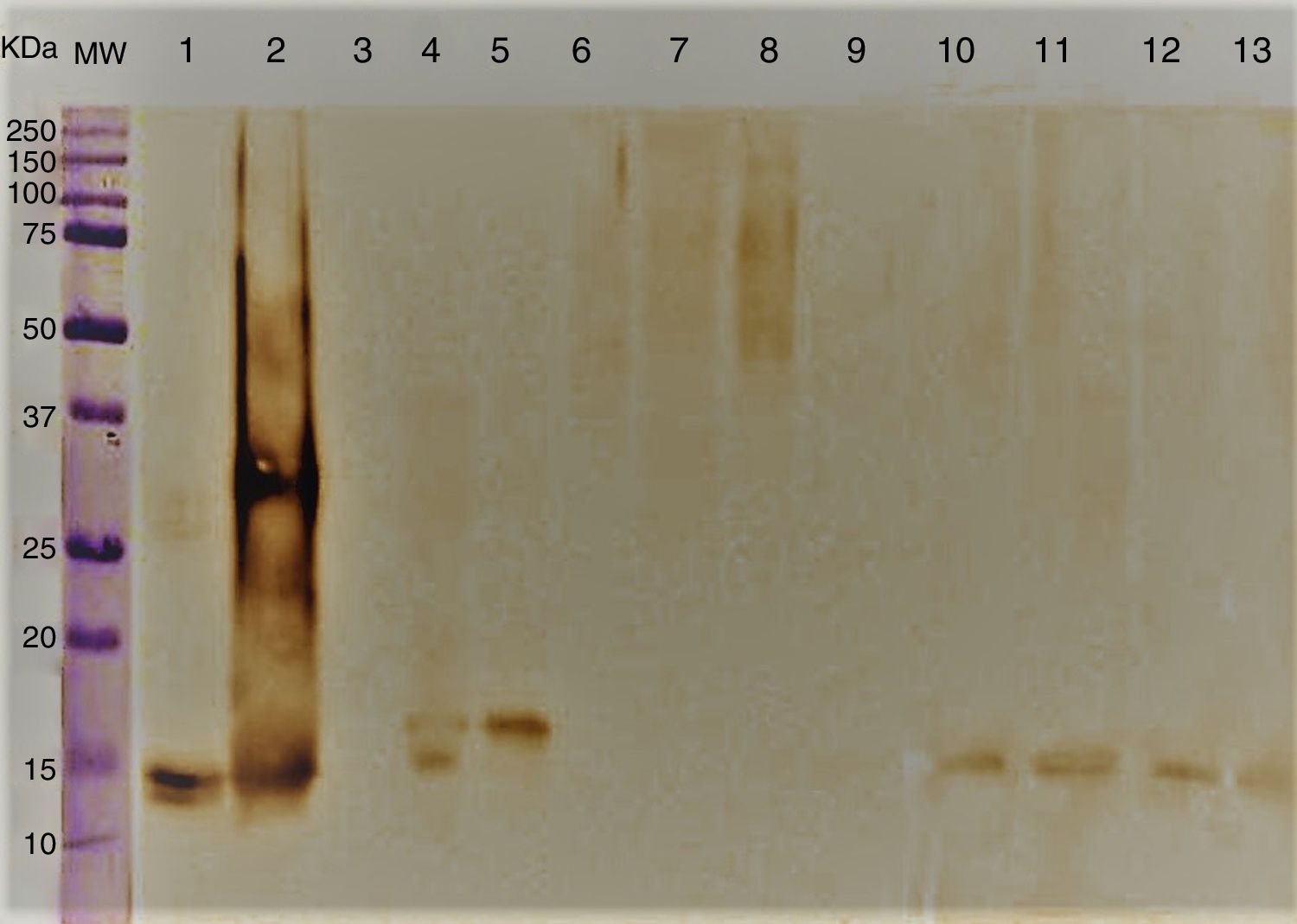
Immunoblotting results showed antigenic components ranging between 14 and 16kDa corresponding to A. alternata PTCC 5224 and airborne strains S. botryosum and U. chartarum which strongly reacted to specific anti rAlt a 1-IgG. However, C. cladosporioides, P. chrysogenum, A. fumigatus, E. nigrum and C. lunata did not show any immunological reaction (Fig. 2).
Immunoblotting of A. alternata and related genera; molecular weight marker protein (1), A. alternata (2), rAlt a 1 protein (3), E. nigrum (4), S. botryosum (5), U. chartarum (6), C. lunata (7), C. cladosporioides (8), A. fumigatus AF-54 (9), P. chrysogenum(10) and A. alternata airborne strains (11–14).
PCR products were analyzed in agarose gel electrophoresis. In all A. alternata strains, U. chartarum, and S. botryosum a 129 bp-fragment was amplified, while in C. cladosporioides, E. nigrum, C. lunata, and A. fumigatus AF-54 this fragment was not present. The amplification of Alt a 1 was confirmed by sequencing and Blast analysis (http://www.ncbi.nlm.nih.gov/BLAST). The polymorphic sites of the amplified gene were compared among A. alternata, U. chartarum, and S. botryosum using MEGA version 6.0 (Fig. 3).
Real-time PCRAlt a1 gene was expressed at very high basal levels in A. alternata PTCC 5224, the same as A. alternata airborne isolates, while it was expressed at low basal levels in S. botryosum and U. chartarum. No significant gene expression was found for C. lunata. With regard to Alt a 1 encoding gene expression, the Ct mean value for A. alternata strains ranged from 24.70 to 27.84, while for the related taxa this value ranged from 23.62 to 32.09.
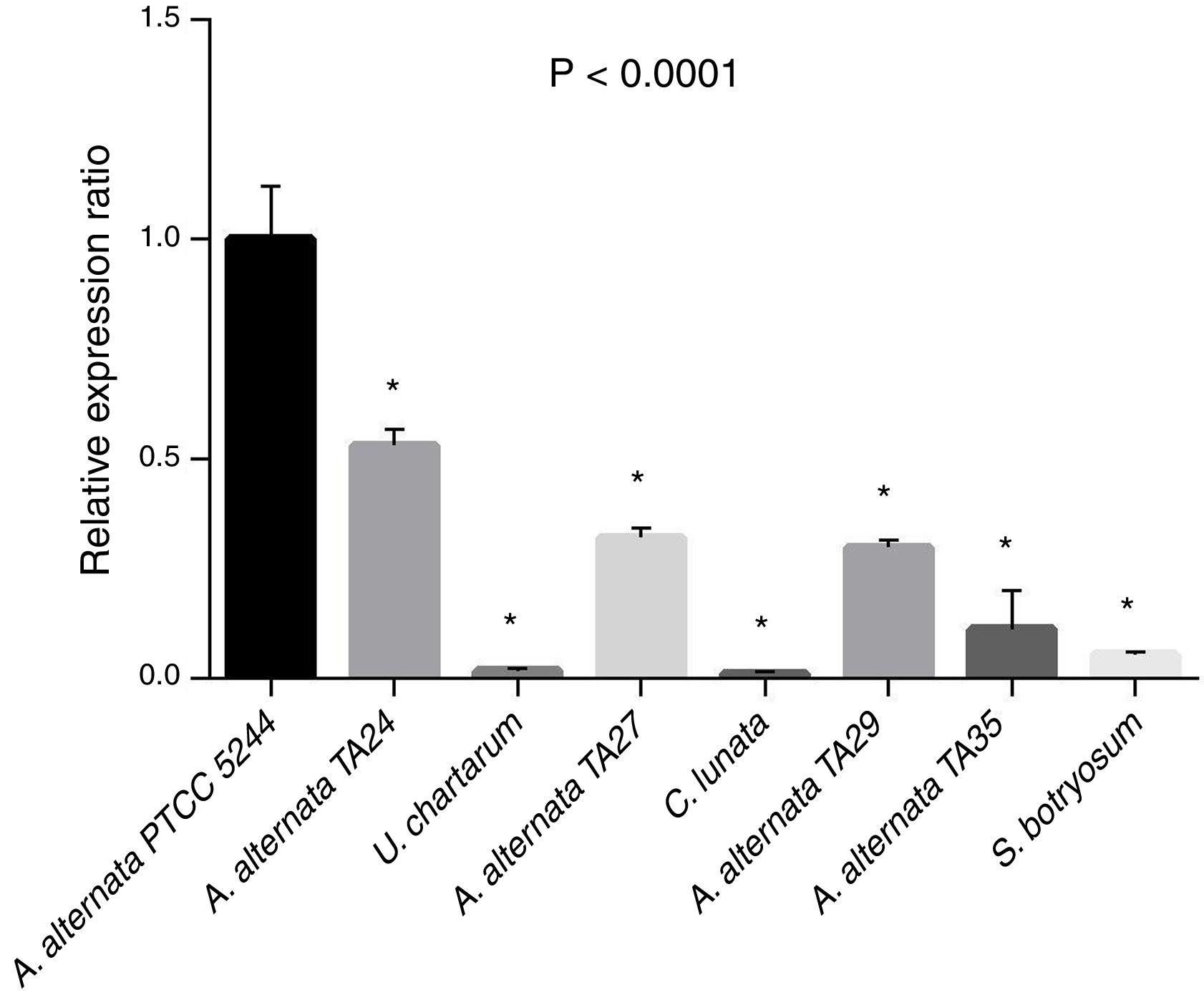
The relative expression ratio (RQ) results were determined in A. alternata PTCC 5224 (RQ=1), airborne A. alternata TA24, TA27, TA29, and TA35 (RQs=0.53, 0.32, 0.29, 0.11, respectively), U. chartarum (RQ=0.05), S. botryosum (RQ=0.016), and C. lunata (RQ=0.01) (Fig. 4). The highest RQ was found in A. alternata PTCC 5224 and the expression level was declined in airborne A. alternata strains by 50–90%, in Ulocladium chartarum by 95% and in S. botryosum by 98%. Curvularia lunata showed the least expression among the tested fungi.
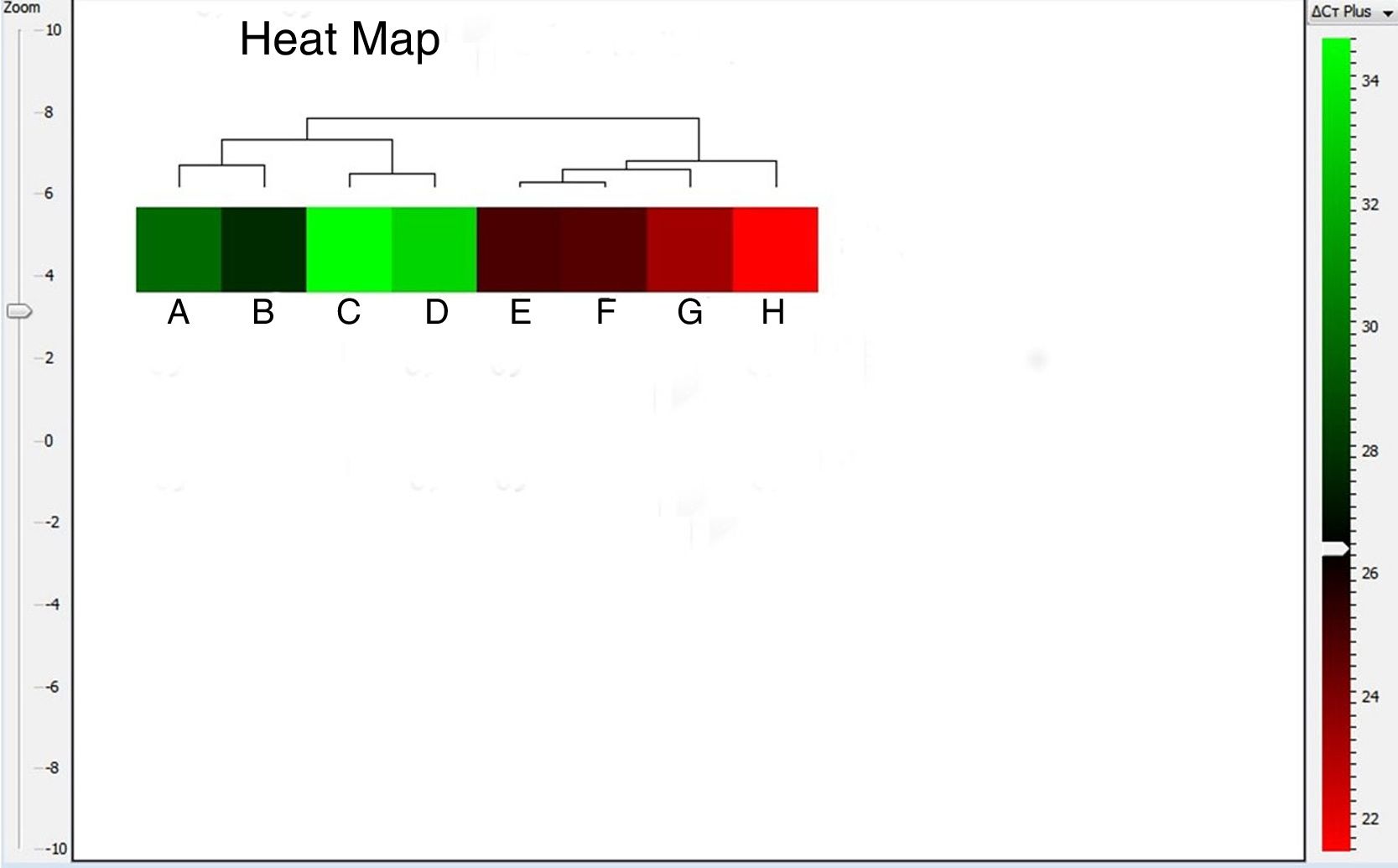
The heat map can be configured as either red-blue or red-green map, where the red color box represents up-regulated gene expression. As shown in Fig. 5, A. alternata PTCC 5224 and airborne Alternaria isolates showed the highest levels of Alt a 1 expression. The U. chartarum and S. botryosum strains revealed an intermediate level of Alt a 1 expression while C. lunata showed very low expression level of this gene. The dendrogram and heat map indicate that Alt a 1 expression is well correlated with the presence of the allergenic Alt a 1 protein in A. alternata, showing the importance of this allergenic protein in other related genera and species of the Pleosporaceae family.
DiscussionThe complexity of the allergenic compounds of A. alternata makes it difficult to establish a relationship between rhinitis and/or asthma symptoms in allergic individuals sensitized to A. alternata or to other allergens different from Alt a 1.13,38 It is well known that while A. alternata shows limited cross-reactivity with allergenic molds, such as A. fumigatus, P. chrysogenum and C. cladosporioides, it has significant levels of allergenic cross-reactivity with other fungi in the Pleosporaceae family.18,27 In the present study, culture filtrates of standard and airborne isolates of A. alternata, S. botryosum, and U. chartarum showed strong reactivity with specific serum anti rAlt a 1. Nevertheless, we did not find any reaction in C. lunata, C. cladosporioides, and A. fumigatus culture filtrates, thus indicating the absence of Alt a 1 homologs in these species.6,30 Variability could be related to the contents and potency of A. alternata secreted allergens.3,18,23,27 Duffort et al.12 found that fungal extracts obtained from Cladosporium or Aspergillus could lack structures similar to Alt a 1. Highly specific protein for Alt a 1 was found in culture filtrate extracts of Stemphylium, which belongs to the same family as A. alternata (Pleosporaceae family).3,17 Also, Moreno et al.24 suggested that the allergen Alt a 1 of A. alternata could be considered a non-species-specific allergen that could be used as a diagnostic source of sensitization to some species of the Pleosporaceae family.
The real-time PCR assay described in the present study showed a 100-fold higher sensitivity than that of conventional PCR techniques. Our results revealed the presence of the major allergen Alt a 1 in A. alternata and other related fungi in the Pleosporaceae family. Furthermore, a quantification of Alt a 1 in the culture filtrates of A. alternata and airborne Alternaria isolates showed the same significant amount of the encoding gene expression. It was shown that S. botryosum and U. chartarum expressed the Alt a 1 gene moderately, while C. lunata revealed no gene expression in qRT-PCR. These results further indicate that qRT-PCR could be a useful tool to quantify the major fungal allergens (Alt a 1) causing allergic diseases. Targeting the Alt a 1 gene has been developed for the rapid detection of DNA by PCR amplification for phylogenetic analysis of Alternaria and related genera.15,18,29,37 Rosenthal et al.29 found that mRNA encoding Alt a 1 protein was present in eight different strains of A. alternata, with similar concentrations in seven of the eight strains. Hong et al.18 amplified chromosomal genes homologous to Alt a 1 in species of Alternaria, Stemphylium, Ulocladium, Nimbya and Embellisia.18 They showed that despite high sequence variation of Alt a 1, secondary structure was predicted to be well preserved in most of the species. Their findings suggested that Alt a 1 gene is unique to specific groups of fungi including Alternaria and related species. However, it remains very important to examine whether Alt a 1 homologs from diverse Pleosporaceae members have cross-reactivity with IgE against specific Alt a 1 despite variation in primary and secondary structures.18,30 It has been demonstrated that culture filtrate extracts from S. botryosum and U. botrytis contain antigenic proteins that react with IgE antibodies specific to rAlt a 1 from experimental serum.17 Alt a 1 has been detected in the atmosphere in a concentration that correlated well with mold spore counts and with the total Alternaria allergen activity in the atmosphere.2,14,28 For Postigo et al.27 Alt a 1 can be a promising candidate for a molecular-based approach to the diagnosis and therapy of A. alternata. Allergic reaction to fungi as a cause of these symptoms has not been established by appropriate skin testing or in vitro IgE tests. Standardization, prevention, diagnosis and therapy of allergic diseases have contributed to the characterization and identification of the individual components causing allergic diseases and their possible biological and immunological relationships.10 Further evaluation of the Alt a 1 expression by Real time-PCR in our study showed the different levels of gene expression among the strains and species. A. alternata standard strain showed the highest level of Alt a 1 expression. While the gene expression levels of A. alternata airborne isolates were similar to the standard strain, the gene expression was moderate in U. chartarum and S. botryosum, and very limited in C. lunata.
In this study we checked standard strains of different important genera of the Pleosporaceae family to find out whether Alt a 1 gene is expressed in genera other than Alternaria. Taken together, our findings have shown the presence of Alt a 1 and an efficient expression of Alt a 1-related encoding gene in A. alternata and some other important genera of the Pleosporaceae family such as U. chartarum and S. botryosum. Despite there is no data regarding host–pathogen interaction in our study, our findings suggest that Alt a 1 could play a preponderant role in the etiology of fungal allergy and asthma in those individuals susceptible to this allergen. Overall, the study of Alt a 1 homology in A. alternata and other members of the Pleosporaceae family and the evaluation of its role in the etiology of allergy in asthmatic patients and other susceptible individuals is highly recommended.
Conflict of interestThe authors declare that there is no conflict of interest relevant to this article.
This study was financially supported by the Research Deputy of Tarbiat Modares University.