In a previous work we showed the feasibility of an interferon gamma release assay (IGRA) for detecting latent infection by Histoplasma capsulatum. While in that proof-of-concept study we used crude fungal extracts as antigens, the newest IGRAs developed for other infections are based on molecularly defined antigens, mostly on mixtures of immunogenic peptides.
AimsTo identify proteins in H. capsulatum that might serve as molecularly defined antigens for an IGRA test.
MethodsWe surveyed the literature looking for known H. capsulatum-immunogenic proteins and assayed two of them as antigens in an IGRA test, in a study that involved 80 volunteers. Furthermore, we used several bioinformatics tools to identify specific H. capsulatum proteins and to analyze possible strategies for the design of H. capsulatum-specific immunogenic peptides.
ResultsSeven H. capsulatum-immunogenic proteins were retrieved from the literature. IGRA tests using either the heat shock protein 60 or the M antigen showed high sensitivities but low specificities, most likely due to the high sequence similarity with the corresponding orthologs in other pathogenic microorganisms. We identified around 2000 H. capsulatum-specific proteins, most of which remain unannotated. Class II T-cell epitope predictions for a small number of these proteins showed a great variability among different alleles, prompting for a “brute force” approach for peptide design.
ConclusionsThe H. capsulatum genome encodes a large number of distinctive proteins, which represent a valuable source of potential specific antigens for an IGRA test. Among them, the Cfp4 protein stands out as a very attractive candidate.
En un trabajo anterior mostramos la viabilidad de un ensayo de liberación de interferón-gamma (IGRA) para detectar la infección latente por Histoplasma capsulatum. En esa prueba de concepto utilizamos extractos crudos del hongo como antígenos; sin embargo, los IGRA de última generación desarrollados para otras infecciones se basan en antígenos definidos molecularmente, principalmente en mezclas de péptidos inmunogénicos.
ObjetivosIdentificar proteínas de H. capsulatum que podrían servir como antígenos definidos molecularmente en una prueba IGRA.
MétodosExaminamos la literatura en busca de proteínas inmunogénicas de H. capsulatum ya conocidas, y ensayamos dos de ellas como antígenos en una prueba IGRA, en un estudio donde participaron 80 voluntarios. Además, utilizamos varias herramientas bioinformáticas para identificar proteínas específicas de H. capsulatum y analizar posibles estrategias para el diseño de péptidos inmunogénicos específicos.
ResultadosEncontramos siete proteínas de H. capsulatum caracterizadas como inmunogénicas en la literatura. Las pruebas IGRA donde utilizamos la proteína de choque térmico 60 o el antígeno M, mostraron una alta sensibilidad, pero baja especificidad, debido probablemente a la alta similitud de secuencia con los ortólogos correspondientes en otros microorganismos patógenos. Identificamos unas 2000 proteínas específicas de H. capsulatum, la mayoría de las cuales permanecen sin anotar. Las predicciones de epítopos de células T de clase II realizadas para un pequeño número de estas proteínas mostraron una gran variabilidad entre los diferentes alelos, sustentando la aplicación de un enfoque de «fuerza bruta» en el diseño de estos péptidos.
ConclusionesEl genoma de H. capsulatum codifica una gran cantidad de proteínas específicas que representan una fuente valiosa de posibles antígenos para una prueba IGRA. Entre ellos, la proteína Cfp4 resulta un candidato muy atractivo.
Histoplasmosis, caused by the dimorphic ascomycete fungus Histoplasma capsulatum, is the most common endemic mycosis in the Americas.9 Because immunocompromised individuals have a high risk of developing a disseminated H. capsulatum infection, the HIV pandemic and the use of immunosuppressive drugs have led to a dramatic increase in lethal histoplasmosis,2,23,28 developed either from primary infections and the re-burst of old ones with latent H. capsulatum foci.25 Currently, no laboratory test can efficiently detect sub-clinical histoplasmosis, which may remain in a latent form even after several years from the primary infection.
In a previous proof-of-concept study24 we showed the feasibility of an interferon gamma release assay (IGRA) specific for H. capsulatum to detect latent infection in asymptomatic individuals who suffered a H. capsulatum infection in the recent or distant past (from several months up to many years ago). The antigens tested in that first study were two histoplasmins and two yeast filtrates from different H. capsulatum strains (crude preparations).
In spite of the encouraging results obtained with the H. capsulatum-antigen crudes,24 using these antigens may not be a suitable option for the development of a commercial IGRA test. Indeed, a survey of the literature shows that the different IGRA tests developed for several infection models have evolved from the use of crude antigens to the identification of unique proteins in the microorganism of interest and the use of peptide mixtures derived from these proteins. A relevant example of this is the evolution of QuantiFERON-TB for Mycobacterium tuberculosis (Mtb). Its first version was based on the same purified protein derivative (PPD) used in the traditional tuberculosis skin test (TST), whereas the next version (QuantiFERON-TB Gold) used a mixture of synthetic, overlapping peptides that fully cover the sequences of two M. tuberculosis-specific proteins (ESAT-6 and CFP-10), designed to be presented by class II HLA molecules and therefore being recognized by T-CD4+ lymphocytes.1,22
Peptide mixtures following different conceptual designs have been used in several other IGRA tests targeting different infections, as cytomegalovirus (QuantiFERON-CMV),13Leishmania,12,27 hepatitis B virus,8 varicella-zoster virus,26Toxoplasma gondii,7Mycobacterium leprae29 and HIV.14 Purified or recombinant proteins have been also used, as in Leishmania (histone B2 protein),27M. leprae (18, 65 and 70kDa purified antigens)29 and HIV-1 (p24 and gp120 recombinant proteins).16
In this work, using information from the literature, bioinformatics tools and assaying molecular candidates in an IGRA test that involved dozens of volunteers, we sought to identify proteins from H. capsulatum that might serve as molecularly defined antigens for a H. capsulatum-specific IGRA test, either in the form of whole recombinant antigens or as mixtures of synthetic peptides designed from their amino acid sequences.
Materials and methodsIdentification of H. capsulatum antigensTwo alternative strategies were explored in parallel to identify possible molecularly defined antigens that might be used in a H. capsulatum-specific IGRA test: (i) Searching for H. capsulatum proteins that have been previously characterized as immunogenic molecules; and (ii) Identification of specific H. capsulatum proteins, that is, proteins that do not have close orthologs in other related fungi. In both cases, the identified proteins might be used in the form of complete proteins or as sources of specific immunogenic peptides. In the second approach, we used for comparison the sequences of all the proteins identified in the four H. capsulatum genomes that have been sequenced so far: G186AR, H88, H143 and NAm1, having 9315; 9449; 9582 and 18,736 identified proteins, respectively. Comparisons were made against the proteomes of four phylogenetically close fungal species: Blastomyces dermatiditis (11,211 proteins), Emmonsia parva (8562), Paracoccidioides brasiliensis (16,834) and Paracoccidioides lutzii (18,015). The identification of groups of orthologous proteins in these proteomes was carried out with the OrthoMCL program.21 Specific H. capsulatum proteins were analyzed with the TMHMM prediction method20 to predict transmembrane domains; SignalP 3.0 and SecretomeP6 were used to predict signal peptides and secretion proteins, respectively, and TargetP11 was used to predict mitochondrial proteins.
Sequences analyses and epitope predictionThe NCBI BLAST server (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to compare the sequences of selected H. capsulatum proteins against the non-redundant NCBI sequence database. Predictions of class II epitopes were performed with the NetMHCII 2.3 server.19
Recombinant H. capsulatum antigensThe M antigen and the heat shock protein 60 (HSP60), both fused to a C-terminal His tag, were produced from previously obtained Escherichia coli clones.14,15 The proteins were purified by metal chelate chromatography using a Ni2+-Sepharose affinity column (TALON® Metal Affinity Resins; Clontech Laboratories Inc., USA). Protein yield was verified by SDS-PAGE, as well as by Western blot, using the serum from a H. capsulatum-infected individual as the source of primary antibodies against H. capsulatum antigens. Binding of serum antibodies was detected with an alkaline phosphatase-conjugated goat anti-human IgG antibody (Sigma-Aldrich, USA). For both antigens, thick bands corresponding to the expected molecular weights (between 72–95kDa and ∼60kDa for the M antigen and HSP60, respectively) were observed.
IGRA testsThe study was conducted at the Corporación para Investigaciones Biológicas (CIB) in Medellin, Colombia. Eighty individuals who participated in our previous proof-of-concept study24 volunteered for this second exploratory study. These were immunocompetent individuals with negative HIV test and age between 10 and 82 years. Pregnancy and immunosuppressive conditions were exclusive criteria. All the participants provided a written informed consent after receiving a thorough description of the study. Eighteen out of the 80 individuals never, presumably, had contact with H. capsulatum and were assigned to Group A (non H. capsulatum-infected), whereas 11 individuals (Group B-proven) showed clinical records proving that they were infected with H. capsulatum in the past. The remaining 51 individuals showed risk factors of exposure to H. capsulatum (Group B-risk). The assay was conducted as described by Rubio-Carrasquilla et al.24 Briefly, whole blood (500μl/well) was stimulated with 10μg/ml of either HSP60 or M antigen. Non-stimulated blood-wells (basal level IFN-γ production) and blood-wells stimulated with 20μg/ml of the T cell mitogen phytohemagglutinin (PHA) (Sigma-Aldrich, USA) were used as negative and positive controls, respectively. Cultures were incubated at 37°C with 5% CO2 for 24h and then supernatants were collected. IFN-γ concentration in the supernatant was measured using a commercially available ELISA kit (Duo-set R&D Systems, Minneapolis, USA), following manufacturer's instructions. Optical density was measured with an ELISA microplate reader (Bio-Rad Laboratories Inc., USA) at 450nm.
Statistical analysesStatistical analyses were performed using R-project version 3.4.2. The Shapiro–Wilk normality test was employed to analyze the empirical distribution of the IFN-γ data, while the Levene's test was used to assess homoscedasticity. The Kruskal–Wallis non-parametric test and the post hoc Dunn's test were used for multiple comparisons of IFN-γ medians between the groups. A Benjamini–Hochberg correction was applied to control the type I error and p-adjusted (q) values were calculated. The assay sensitivity was evaluated as the percentage of positive responses in B-proven group, while specificity was calculated as the percentage of negative responses in the control group A. Statistical differences were considered significant if p-adjusted (q) was less than 0.05.
Ethics statementAll study procedures and written consent forms were approved by CIB's ethics review board.
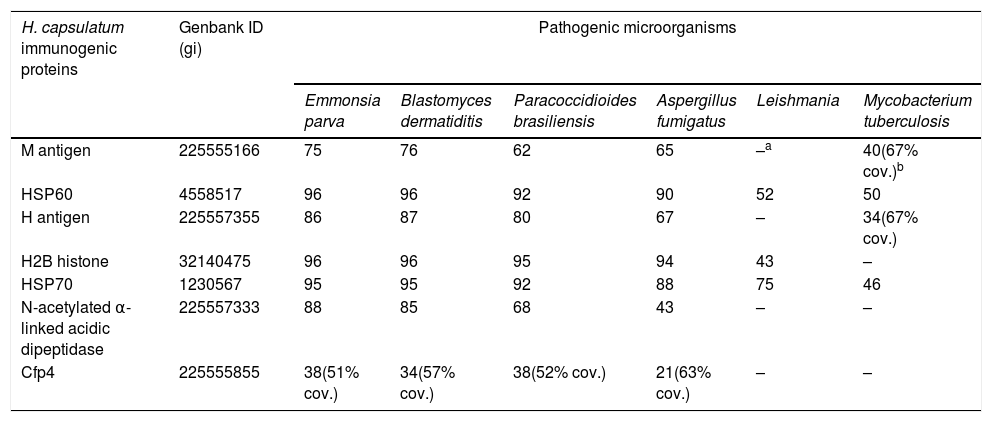
Results and discussionImmunogenic H. capsulatum proteins known from the literatureTable 1 lists seven proteins which have been evaluated before in multiple studies aiming to establish their role in the pathogenesis and immune response against H. capsulatum. Some of these proteins are part of the membrane and cell wall of the yeast phase, while others are secreted to the extracellular environment. Because of their proven immunogenicity, these proteins represent possible molecular antigens for a H. capsulatum-specific IGRA test.
Sequence similarity (given as percentage of amino acid identity) among the seven immunogenic H. capsulatum proteins and their orthologs in related fungi and other unrelated microorganisms.
| H. capsulatum immunogenic proteins | Genbank ID (gi) | Pathogenic microorganisms | |||||
|---|---|---|---|---|---|---|---|
| Emmonsia parva | Blastomyces dermatiditis | Paracoccidioides brasiliensis | Aspergillus fumigatus | Leishmania | Mycobacterium tuberculosis | ||
| M antigen | 225555166 | 75 | 76 | 62 | 65 | –a | 40(67% cov.)b |
| HSP60 | 4558517 | 96 | 96 | 92 | 90 | 52 | 50 |
| H antigen | 225557355 | 86 | 87 | 80 | 67 | – | 34(67% cov.) |
| H2B histone | 32140475 | 96 | 96 | 95 | 94 | 43 | – |
| HSP70 | 1230567 | 95 | 95 | 92 | 88 | 75 | 46 |
| N-acetylated α-linked acidic dipeptidase | 225557333 | 88 | 85 | 68 | 43 | – | – |
| Cfp4 | 225555855 | 38(51% cov.) | 34(57% cov.) | 38(52% cov.) | 21(63% cov.) | – | – |
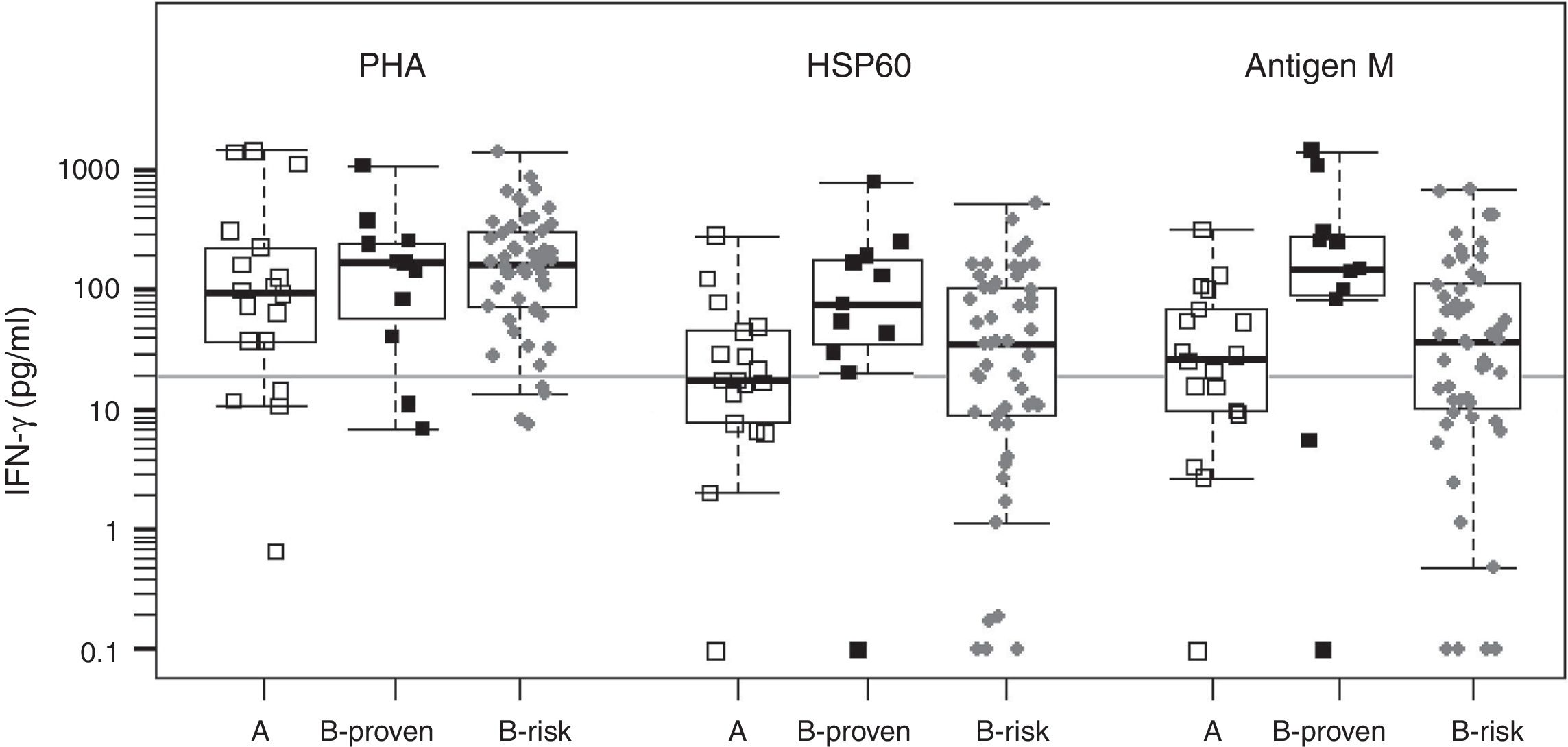
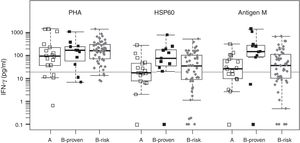
We evaluated two of the H. capsulatum immunogenic proteins, HSP60 and M antigen, both in the form of recombinant proteins, as antigens in an IGRA test. The basal IFN-γ levels, in non-stimulated wells, were within a range of 0–36.8pg/ml, with a median value of zero (data not shown). Fig. 1 shows the IFN-γ responses by group for each antigen. The two recombinant antigens were able to stimulate strong IFN-γ responses in the three groups. The response levels obtained for groups A (non H. capsulatum-infected) and B-proven show a strong overlapping for both antigens. For this reason, it is not possible to define a meaningful IFN-γ concentration cutoff value to discriminate between positive and negative responses. On the other hand we lack a gold standard to compare with, since none of the currently available tests is able to detect sub-clinical H. capsulatum infections. Therefore, we decided to use the cutoff value defined in our previous work, based on the IFN-γ responses to H. capsulatum crude antigens (20pg/ml).24
Statistical box plots for IFN-γ production upon stimulation with PHA and the recombinant proteins HSP60 and antigen M. Symbols
, , represent individuals in groups A, B-proven and B-risk, respectively. The vertical axis is in logarithmic scale. For each individual, the IFN-γ response was adjusted by subtracting the corresponding basal level. IFN-γ concentrations between 0 and 0.1pg/ml were assigned a value of 0.1pg/ml to fit into the plot. Since the data have no normal distribution and are not homoscedastic, the Kruskal–Wallis non-parametric test and the post hoc Dunn's test were used for multiple comparisons of the medians (shown as thick bars within the boxes). The gray horizontal line is placed at the defined IFN-γ cutoff level (20pg/ml).As expected from the strong overlapping of the IFN-γ responses, no statistically significant differences were found between groups A and B-proven for any of the antigens. Using the defined IFN-γ concentration cutoff, the obtained sensitivity values were high (91% and 81% for HSP60 and the M antigen, respectively), with very high IFN-γ response levels to the M antigen, in particular in the B-proven group. However, the specificities were low for both recombinant antigens (56% and 39%, respectively), with several members of group A (non H. capsulatum-infected) showing a medium-level or even a high IFN-γ response.
In B-risk group, 57% (29/51, for HSP60) or 63% (32/51, for M antigen) of the individuals were classified as infected using the above defined cutoff value. The percentage of positive responses in this group is much higher than the one obtained with the H. capsulatum crude antigens,24 most likely because of the lower specificity shown by the two recombinant antigens. All the individuals in B-risk group had been classified as negative by the serology tests.24 Thus, and in spite of its low specificity, the IGRA test performed here may be revealing a latent infection in a number of these individuals, especially in those yielding high (>100pg/ml) IFN-γ responses.
It is worth recalling that M antigen (together with H antigen) is a standard protein tested by serology to detect H. capsulatum infection. Furthermore, both the M antigen and HSP60 are secreted by the fungus3 and therefore are components of the H. capsulatum crudes used as antigens in our first IGRA design, which showed a high specificity.24 Possibly, using these antigens in pure form and, therefore, in greater quantities when compared to their fractions in the crude H. capsulatum preparations, should produce a higher level of antigen presentation, which might cause the exposure of epitopes that are cryptic in H. capsulatum, but dominant in other microorganisms.10 The results obtained indicate that neither HSP60 nor M antigen are suitable antigens for a H. capsulatum-specific IGRA test.
High sequence similarity between H. capsulatum immunogenic antigens and their orthologs in other pathogensThe sequences of the seven immunogenic H. capsulatum proteins were compared against those deposited in Genbank. Six of these antigens showed a high similarity with their orthologs in related fungi (Table 1). The two heat shock proteins (HSP60 and HSP70), in addition, have similar orthologs in other less related pathogens such as Leishmania and M. tuberculosis (Table 1), as well as in other microorganisms (not shown). It is then plausible that the high IFN-γ responses produced by HSP60 and the M antigen in a significant number of non-H. capsulatum infected individuals are most likely cross reactions caused by the immune memory developed by these individuals against other infections. Furthermore, these results suggest that none of the first six proteins from Table 1 are suitable for an IGRA test, at least in the form of whole antigens.
Cfp4 is a H. capsulatum-specific proteinIn contrast with the other six immunogenic proteins, Cfp4 has no close orthologs in related fungi. Furthermore, a BLAST search showed that there are no proteins similar to Cfp4 in any other organism (results not shown). In addition to its high specificity, Cfp4 is very attractive as a possible antigen for a H. capsulatum-specific IGRA test because of two other reasons: it is an immunogenic protein abundantly expressed and secreted in the yeast phase of the fungus17 and has a relatively small size, which allows covering its entire sequence with a limited number of overlapping peptides.
Prediction of H. capsulatum-specific proteinsWe searched for proteins that are distinctive of H. capsulatum by comparing the sequences of all H. capsulatum proteins with the proteomes of other four related species of fungi, as detailed in Methods. The calculations performed with OrthoMCL yielded 11,344 homolog groups, out of which 1983 groups were composed only by homologous proteins from the four H. capsulatum strains. Since it is not practical to analyze each of these proteins individually, we focused on annotated proteins (that is, those with known or predicted function). This resulted in only 41 proteins. The next filter aimed to identify which of these proteins are secreted to the external medium. Using the SignalP server, we obtained five proteins that could be released by a classical secretion route. The remaining 36 proteins were processed with the SecretomeP server, which predicted 13 proteins associated with non-classical secretory pathways. The resulting 18 secretion proteins were further analyzed with the TargetP server, which indicated that none of them are mitochondrial proteins. These 18 proteins were also analyzed with the TMHMM server, which predicted the presence of transmembrane helices in six of these proteins, that were consequently discarded.
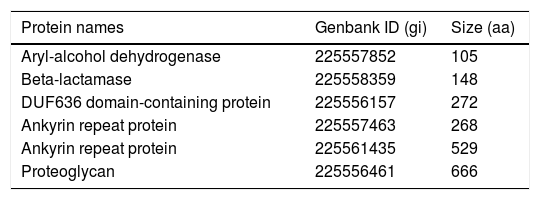
BLAST server, the last filter, was used to compare the sequence of each of the remaining 12 proteins against the whole NCBI database of non-redundant proteins, to account for possible similarities with proteins from other pathogens, in addition to the four fungal species analyzed with OrthoMCL. Six out of the 12 proteins were discarded because of their significant sequence similarity (defined here as >50% amino acid identity in at least 70% coverage of the target sequence) with proteins from other pathogenic fungi, including species of Emmonsia, Blastomyces, Sporothrix, Aspergillus, Fusarium, Trichophyton, and Coccidioides, among others. Table 2 lists the six selected H. capsulatum-specific secretion proteins.
H. capsulatum-specific proteins selected from the orthology analysis.
| Protein names | Genbank ID (gi) | Size (aa) |
|---|---|---|
| Aryl-alcohol dehydrogenase | 225557852 | 105 |
| Beta-lactamase | 225558359 | 148 |
| DUF636 domain-containing protein | 225556157 | 272 |
| Ankyrin repeat protein | 225557463 | 268 |
| Ankyrin repeat protein | 225561435 | 529 |
| Proteoglycan | 225556461 | 666 |
Cfp4 was not included within the 41 annotated proteins that were selected from the OrthoMCL output. The reason why Cfp4 is not annotated is precisely its lack of similarity with other proteins of known function. Cfp4 was, however, among the approximately 2000 proteins without close orthologs found by the program. Hence, there exists a vast space to be explored for possible specific antigens among the large number of non-annotated proteins found in the H. capsulatum genomes.
Peptide antigens from H. capsulatum-specific proteins: rational design or brute force?As mentioned in the Introduction, the commercial IGRAs available for M. tuberculosis and cytomegalovirus are based on peptide antigens. For M. tuberculosis, although the latest version (QuantiFERON-TB Plus) includes a second tube containing class I T-cell epitopes, the design of the test peptides is still aimed mainly at the stimulation of T-CD4+ lymphocytes.5 For H. capsulatum, a Th1-type response is needed for macrophage activation and efficient yeast clearance.18 In this type of response, T-CD4+ cells are crucial, being the main producers of IFN-γ.4 The T-CD8+ cells also play a role in host defense against histoplasmosis, but with minor consequences when compared to T-CD4+ lymphocytes, in immunocompetent individuals.18 Therefore, a plausible approach toward a future development of a H. capsulatum-specific IGRA is the design of class II peptides from H. capsulatum-specific proteins, such as Cfp4 and the other distinctive proteins identified in this study.
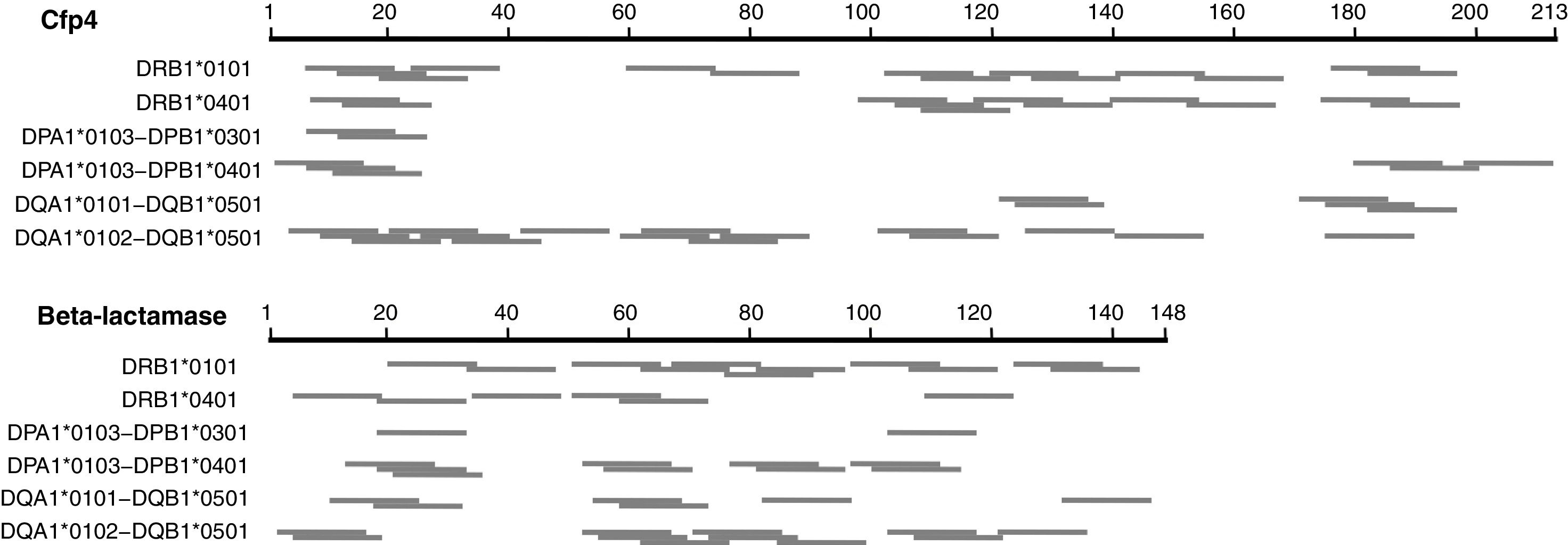
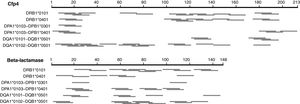
We first explored a rational design approach of class II T-cell peptides. The NetMHCII-2.3 web server was employed to predict possible class II, 15 amino acid long epitopes in the seven distinctive H. capsulatum proteins identified here, for several HLA-DR, DP and DQ alleles serving as models for this exercise. Fig. 2 illustrates the results obtained for Cfp4 and beta-lactamase. Both proteins have a relatively small size, comparable to that of the M. tuberculosis proteins ESAT-6 and CFP-10. It can be observed that the number and distribution of predicted peptides throughout the sequences varies markedly from one allele to another. These results, together with the level of uncertainty that still characterizes the class II prediction algorithms, make the selection of a defined set of peptides (that should cover a wide spectrum of HLA alleles) risky. Taking also into account that the analyzed proteins are relatively short, so that their whole sequences can be covered with a limited number of overlapping peptides, a brute force approach seems more appropriate to create a pool of immunogenic peptides.
Schematic representation of the epitopes predicted with the NetMHCII 2.3 server17 for different class II HLA alleles, for two distinctive proteins of H. capsulatum. The segments in gray represent 15 aa peptides with predicted affinities <500nM. In cases where several consecutive peptides with the same nonapeptide nucleus were predicted, only the central peptide was represented.
The development of an IGRA test for detecting latent infections by H. capsulatum should evolve toward the use of molecularly defined antigens, preferably immunogenic peptides. The use of known immunogenic H. capsulatum proteins having a high sequence similarity with their orthologs in other pathogenic microorganisms may result in a low test specificity, as demonstrated here for M and HSP60 antigens. Our bioinformatics analyses show that the H. capsulatum genome encodes a large number of distinctive proteins, which represent a valuable source of potential specific antigens. The Cfp4 protein, in particular, stands out as a very attractive candidate.
FundingThis work was supported by COLCIENCIAS (grant 221356933526) and the University of Antioquia (CODI-UdeA grant CIEMB-098-13).
Conflict of interestThe authors declare that they have no conflict of interest.