Fusarium oxysporum has worldwide distribution and causes severe vascular wilt or root rot in many plants. Strains are classified into formae speciales based on their high degree of host specificity, of which multilocus sequence typing provides a fairly good estimate.
AimsThe main aim of this study was to identify the causal agent of an infected potato tuber in Colombia.
MethodsTwo F. oxysporum isolates were recovered from a potato tuber showing symptoms of dry rot. Both macroscopic and microscopic morphology differences were observed between the two isolates. Koch's postulates were verified and in quantitative tuber pathogenecity trials, both isolates induced moderate dry rot. Ribosomal internal transcribed spacer (ITS) and partial intergenic spacer region (IGS) sequences were PCR-amplified, sequenced and shown to be identical for the two isolates. A maximum parsimony phylogeny was created using F. oxysporum IGS sequences available in the Genebank database, which does not include sequences from the formae speciales tuberosi.
ResultsOur two isolates were most closely related to a red clover (Trifolium pratense) pathogenic isolate and two non-pathogenic F. oxysporum isolates from birdsfoot trefoil (Lotus corniculatus) and Lycopersicon sp. rhyzosphere (99% identity).
ConclusionsThese experiments showed that our isolates are not restricted to potato and that a molecular marker is needed to differentiate the formae speciales since the IGS and EF-1α do not have the power to do it.
Fusarium oxysporum tiene distribución mundial y causa en varias plantas el marchitamiento vascular severo o la pudrición de la raíz. Las cepas se clasifican en formae speciales basándose en su alto grado de especificidad por un hospedero. Se puede obtener una muy buena estimación de ésta por medio de la tipificación con secuencia de múltiples locus.
ObjetivosEl objetivo principal de este estudio consistió en la identificación del agente causal de un tubérculo infectado en Colombia.
MétodosDos aislados diferentes de F. oxysporum se recuperaron a partir de un tubérculo de papa que mostraba síntomas de pudrición seca. Se observaron diferencias tanto macroscópicas como microscópicas entre los aislamientos. Se verificaron los postulados de Koch y al llevar a cabo ensayos cuantitativos de patogenicidad, ambos aislamientos produjeron los síntomas de la pudrición seca. Se amplificaron por PCR y se secuenciaron regiones del espaciador interno transcrito (ITS) y el espaciador intergénico (IGS) ribosómico.
ResultadosLas secuencias obtenidas fueron idénticas entre los dos aislamientos. Se reconstruyó una filogenia por máxima parsimonia con las secuencias de IGS disponibles en la base de datos de GenBank, que no incluye secuencias de la forma specialis tuberosi. Nuestros dos aislamientos mostraron estar más cercanamente relacionados con un aislamiento patogénico de trébol rojo (Trifolium pratense) y dos aislamientos no patogénicos de Lotus corniculatus y la rizósfera de Lycopersicon sp. (99% identidad).
ConclusionesEstos experimentos demuestran que los aislamientos obtenidos no solo son capaces de afectar a la patata y que se requiere un marcador molecular que permita delimitar las formas especiales ya que ni el IGS ni el EF-1α tienen el poder de resolución suficiente para hacerlo.
Potato dry rot usually occurs during storage and can lead to losses in crop quality and yield.32 Dry rot is caused by several Fusarium species, among which Fusarium sambucinum, Fusarium coeruleum and Fusarium avenaceum are the most common agents in North America and Europe.11,17,30–32 Although it is less frequent and is considered of minor importance in those locations, Fusarium oxysporum f. sp tuberosi is one of the main causal agents of the disease in Tunisia, South Africa, Oregon and Washington.8,14,28,35
The fungus F. oxysporum can be described as a species complex comprising a collection of several clonal lineages.25 It has a worldwide distribution and is responsible for severe vascular wilt or root rot in a wide range of plant families, although non-pathogenic strains are also usually recovered.13,21,25 It is ubiquitous in both agricultural and non agricultural soils, and is generally found in close association with plant roots.40
Strains are classified into more than 150 formae speciales (f. sp) based on their host specificity.13,25 A forma specialis has generally one host, sometimes several, and may also colonize other plant species endophytically.40Formae speciales can be indistinguishable from each other and from non pathogenic isolates, and pathogenicity trials are necessary to identify them with absolute certainty. However, with the use of a combination of molecular markers and vegetative compatibility group tests, isolates can be reasonably associated with a forma specialis in some cases.3,9,16,21,22,24,27,40 Many formae speciales are polyphyletic, comprising several not closely related clonal lineages.21,25,33,35,36,40 This phenomenon can be explained by the emergence of new pathogenic strains by mechanisms such as lateral gene transfer and mutation, however much of this remains to be thoroughly explained.4,19,23,25
Colombia is considered the fourth largest potato producer in Latin America with a production of 1.9 million tons and a cultivated area of 110,000 hectares in 2007.29 Studies on the potato pathogens have mainly focused on diseases such as late blight (Phytophthora infestans),34 bacterial wilt (Ralstonia solanacearum), soft rot (Erwinia spp.) and defects caused by insects and worms.6 Besides, being the first report of F. oxysporum rot in Colombia, the purpose of the present study was to make a preliminary characterization and identification of the isolates obtained from a diseased tuber in Colombia.
Materials and methodsIsolatesIsolates were recovered from potato (Solanum tuberosum sbsp. andigena) tubers using the following procedure. Infected tubers were thoroughly washed, surface-sterilized for 5minutes in 5% (v/v) sodium hypochlorite solution, and rinsed 3 times with sterile distilled water. Then the tubers were sliced using a sterile knife, and incubated at 95% relative humidity and 20°C for 7 days. After incubation, pieces of tissue from the leading margin of lesions, from tuber flesh showing dry rot symptoms, were placed on potato dextrose agar (PDA, Oxoid) amended with 0.5mg/ml chloramphenicol, and incubated for 7 days at 25°C.
Since the recovered isolate showed polymorphism on morphology, single spore cultures were established by serial dilution of a conidial suspension in water with 1% Tween 20 (Sigma), dispersing a drop of 10-6 and 10-8 dilutions on malt agar (MA, Oxoid) and selecting a germinated macroconidium under the microscope, after 24hours incubation at 25°C. Macroscopic characteristics were observed after 7 days of incubation at 25°C in PDA. Microscopic characteristics were observed from cultures in carnation leaf agar (CLA) grown for 14 days at 25°C in the dark.
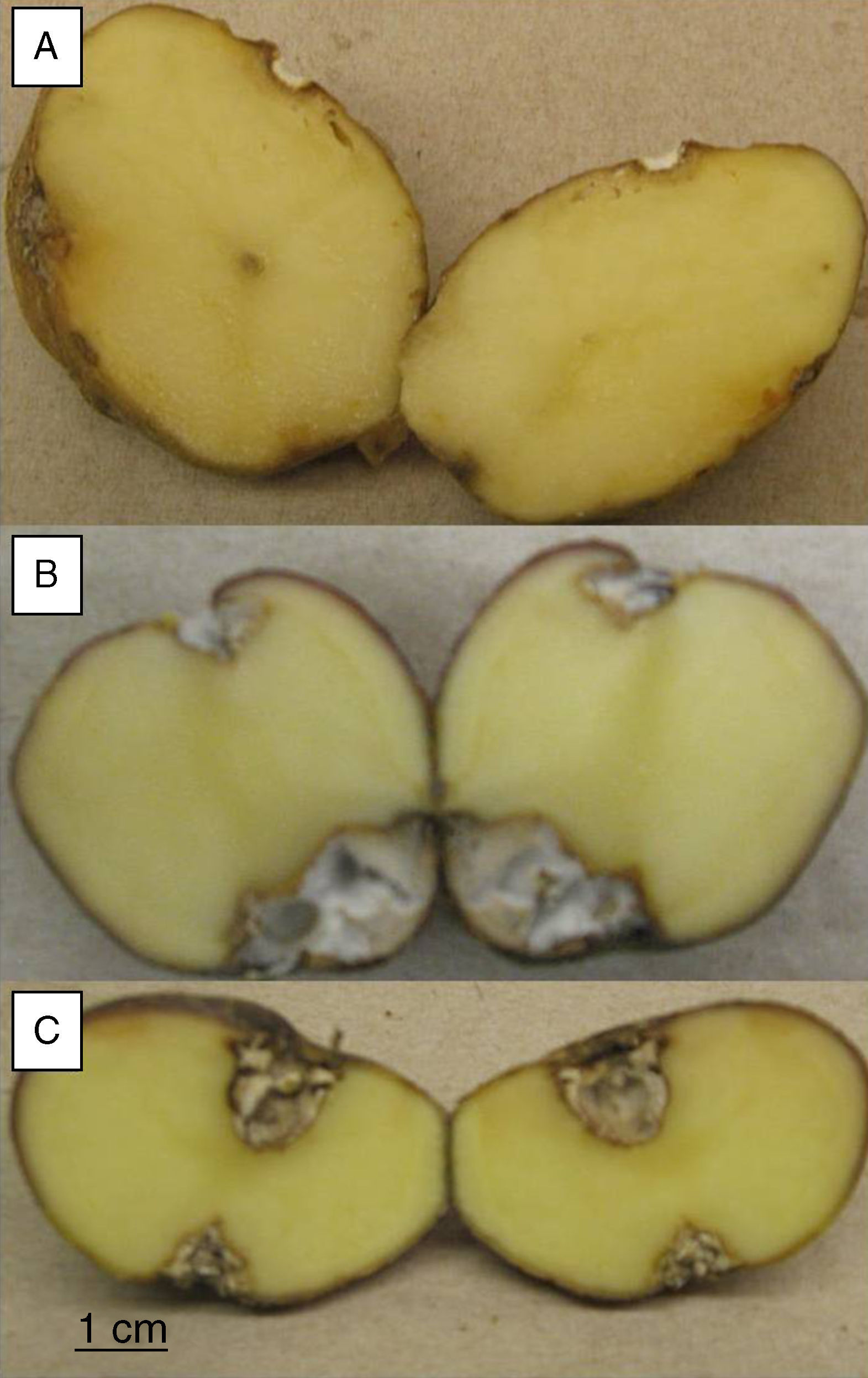
Pathogenicity testsTubers from S. tuberosum sbsp. andigena were washed and surface sterilized by immersion in 5% v/v sodium hypochlorite solution for 15minutes, then rinsed three times in sterile distilled water and air-dried. Two diametrically opposed wounds, 5mm diameter and 5mm deep, were made on each tuber, using a sterile glass rod. A 5-mm-diameter agar plug, from the margin of a 4 day old-culture on PDA, was placed into the wounds and a sterile plug on the control tubers. Tubers were placed in sterile plastic beakers with water soaked paper towels covering the bottom of the beakers, and then were incubated at 25°C in the dark for 30 days. Twelve replicates were made for each strain and the sterile control. After incubation, the tubers were cut across the two inoculation points. Pictures of 2,048 x 1,516 pixels resolution were taken with a Canon Powershot A550 digital camera and the area of the lesions was measured with the image analysis software ImageJ v.1.38 (US National Institutes of Health, Bethesda, Maryland, USA).
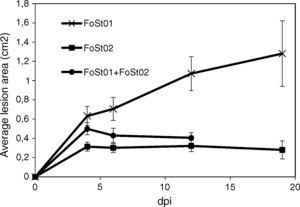
Tuber slice pathogenicity tests (time progression)Tubers were sterilized as described and cut to 15mm width slices. A sterile wooden toothpick was used to scrape a 7 day old fungal culture in PDA, and inoculations were performed by puncturing tuber slices, to about 3mm depth. The slices were put in Petri dishes and incubated at 25°C and 95% RH. Lesion progression was evaluated periodically (at 3, 6, 12 and 19 dpi) for 19 days, as described above. Seven replicates (14 inoculation points) were evaluated for each treatment.
Red clover pathogenicity testsRed clover (Trifolium pratense) seeds were rinsed with distilled water, surface sterilized for 5minutes on 5% ethanol and washed three times with sterile distilled water. Seeds were sown in autoclaved soil, four seeds per 0.5 L of soil. The seedlings were fertilized weekly with a 300ppm solution of Triple 15 fertilizer (15% w/w N, 15% w/w P2O5, 15% w/w K2O). Plants were grown at 20-25°C and watered weekly during four months, when they were harvested. The roots were washed with sterile distilled water and small wounds were induced with a sterile blade. The roots were then submerged in the inoculum prepared for 30minutes before being replanted. Inoculum was obtained by flooding a one week old PDA culture of each fungus with 5ml sterile distilled water and standardizing these inocula to 100,000 microconidia/ml by counting in a Newbauer chamber. Over the next two weeks, the plants were examined for disease symptoms and then harvested to check for signs of root rot. Control plants were submerged in sterile water. In those cases in which the disease was observed, recovery of the pathogen from roots was attempted by washing the roots with 0.5% (v/v) sodium hypochlorite solution for 5minutes, excising and placing the diseased tissue on PDA supplemented with chloramphenicol and incubating for 7 days at 25°C.
Statistical analysesStatistical analyses were implemented in SPSS 16.0 (SPSS Inc. Chicago, Illinois). Normal distribution of all measurements was evaluated with the Kolmogorov-Smirnov normality test and outliers were discarded. For the tuber pathogenicity trials, difference between the means of each treatment were evaluated by a one-way ANOVA, and the Tukey 95% confidence intervals for the means were calculated. For the time progression pathogenicity tests, one-way ANOVA was carried out for each treatment across the different measurements in time. Since no statistically significant differences in the means across time were observed for three of the four treatments, a one-way ANOVA to compare the means between treatments in the first time point was made.
DNA extraction, amplification and sequencingMycelia were obtained by inoculating 4 plugs from a 5-day-old culture on PDA into four 250ml flasks containing 40ml Sabouraud broth (4% glucose, 4% bacto-peptone, 0.5% yeast extract), and then incubating for 4 days at room temperature in a shaker (250rpm). Mycelia were harvested by vacuum filtration through Whatman grade 1 filter paper, and then lyophilized for 24hours before grinding them to fine powder. After grounded, 100mg of the powder were transferred to a 1.5ml Eppendorf tube and DNA was extracted using CTAB, as described by Möller et al. 1992.26 PCR amplification of internal transcribed spacer (ITS) rRNA and partial intergenic spacer (IGS) rRNA regions (245bp to 1,056bp at F. oxysporum IGS region) was performed using the primer pairs ITS3-ITS439 and FIGS11-FIGS12.2,13 Each 25μl reaction contained 1μl DNA, 1 U/μl Fermentas Taq DNA polymerase (Fermentas Life Sciences), 1X Fermentas Taq Buffer, 2.5mM MgCl2, 0.2mM each dNTP, 0.2mM each primer. PCR was conducted in a Bio-Rad MyCyclerTM thermal cycler as described by Appel and Gordon, 1996.2 Amplification products were visualized under UV light on 1% ethidium bromide stained agarose gel. Sequencing was conducted on both strands by Macrogen (Korea). Sequences were deposited in GenBank under accession numbers GU132455 and GU132456.
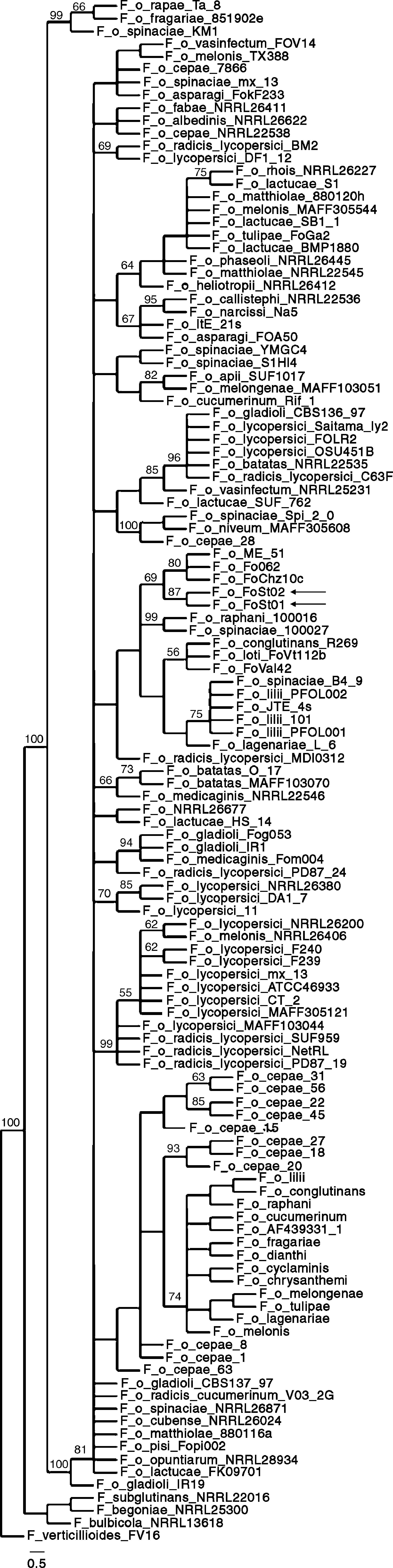
Phylogenetic analysisSimilarity searches (blastn, default parameters) were performed on GenBank with the ITS and IGS sequences. For IGS, the best 100 hits (by score) were retrieved and multiple sequence alignments were done with MUSCLE with default parameters.12 Alignments were manually edited in Jalview 237 to eliminate regions outside the FIGS11-FIGS12 region and discard incomplete sequences. Also, identical sequences from the same formae speciales were discarded to avoid excessive redundancy. Maximum parsimony phylogenies were made with TNT 1.110–15, using traditional search, 1 nucleic acids transversion cost and standard bootstrapping with 1,000 replicates. Retention index, consistency index and number of steps were calculated in PAUP* 4.0 (Sinauer Associates, Inc.). Graphic visualization of the tree was done in TreeDyn.5
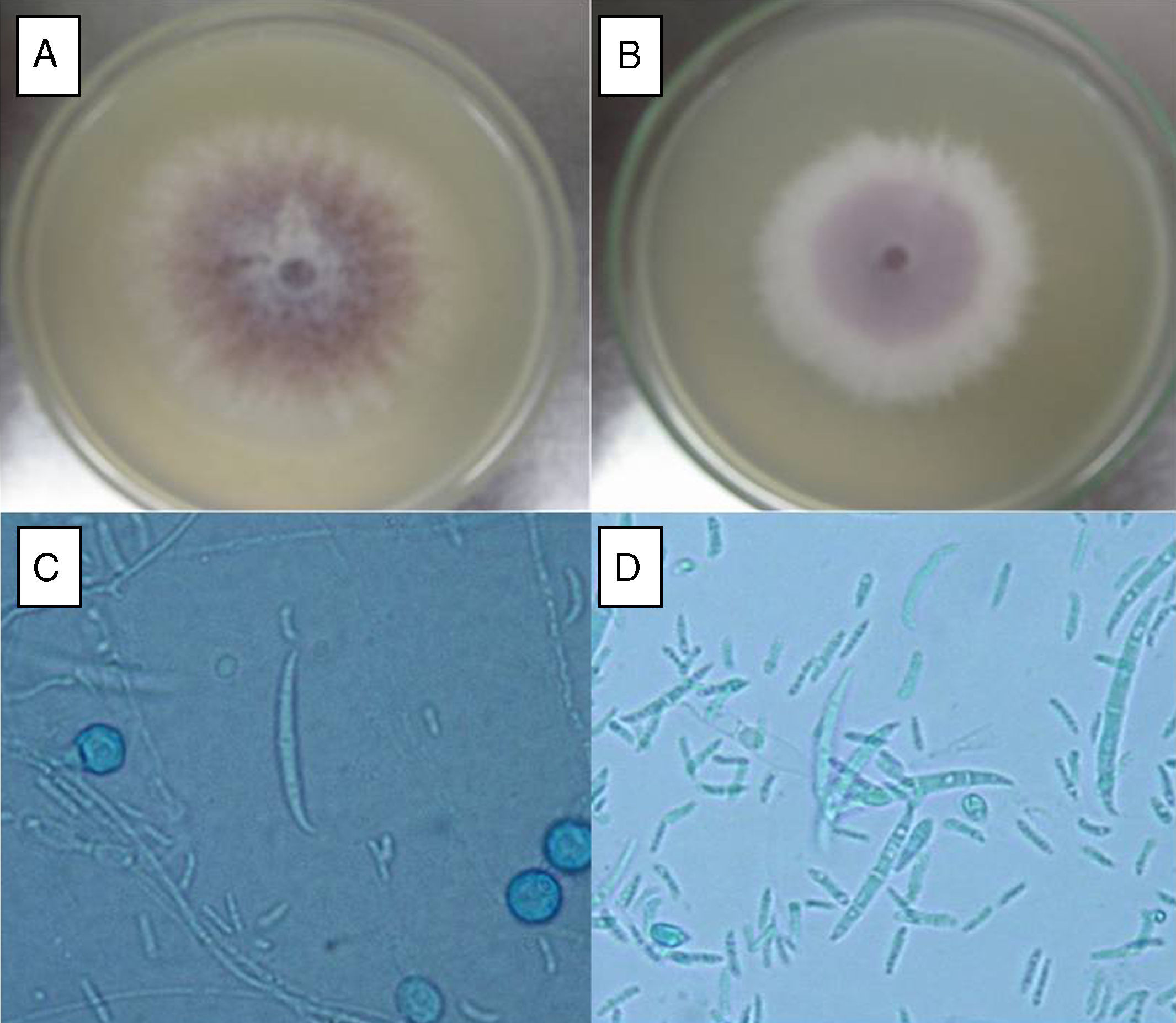
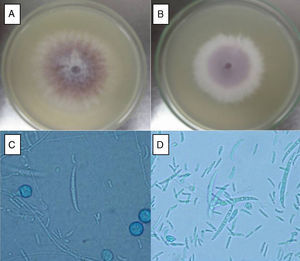
ResultsF. oxysporum isolatesTwo different isolates (FoSt01 and FoSt02) were obtained from a single potato tuber (Solanum tuberosum sbsp. andigena) bought in a marketplace in Bogotá, which showed moderate dry rot symptoms. Given the observation of two different colony morphologies, superposed in the culture, repeated subcultures were performed on malt agar to separate them. Once the isolates were separated, several single spore cultures were obtained. Of these, two distinct colony morphologies were observed, which were named FoSt01 and FoSt02. Even though, both isolates showed macroscopic and microscopic morphologies consistent with the type description for Fusarium oxysporum,20 it was still possible to notice morphological differences between them (Fig. 1A and 1B). On PDA, FoSt01 produced sparse white aerial mycelium with a strong dark purple pigment diffusing into agar, visible from both sides of the Petri dish. Meanwhile, isolate FoSt02 produced abundant light purple aerial mycelium and the underside of colonies was from pale tan to light purple. On CLA, both isolates produced abundant microconidia, which were oval (FoSt02) or kidney shaped (FoSt01) and single or two-celled. They were produced on false heads with short monophialides. Intercalar chlamydospores were abundant, mostly single but sometimes in pairs. Macroconidia were slightly curved, thin walled, generally three septae and with a foot-shaped basal cell (Fig. 1C and 1D).
Pathogenicity testsThe isolates reproduced the symptoms in inoculated tubers and it was possible to recover them back from the inoculated tissue, thus verifying Koch's postulates.
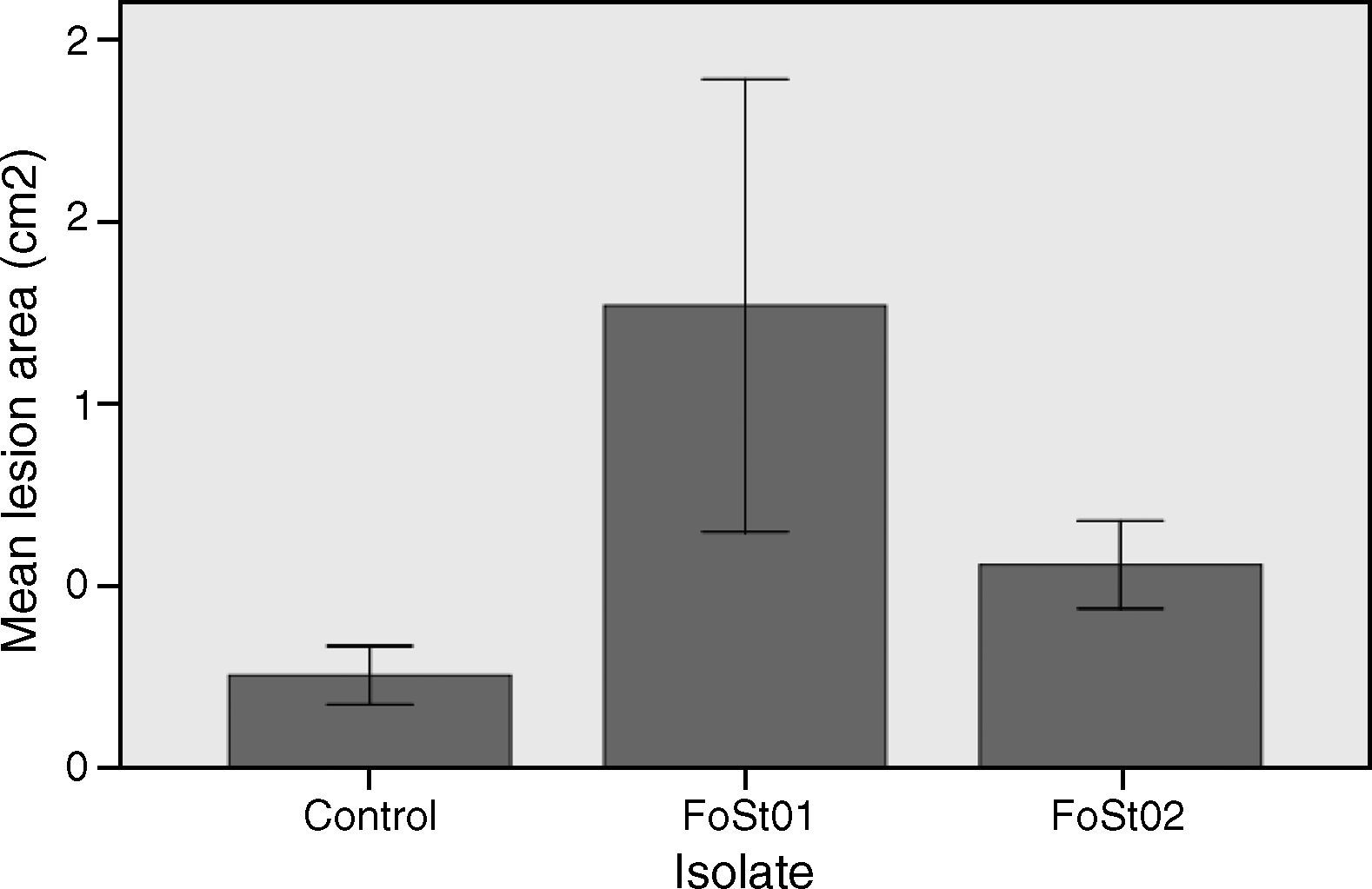
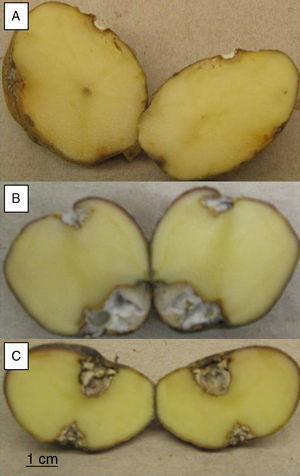
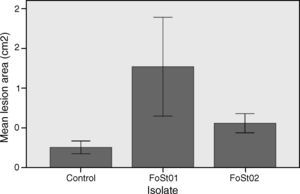
To evaluate the virulence of both isolates, pathogenicity tests on complete tubers were made (Figs. 2 and 3). These showed statistically significant differences among the isolates and with the control (P-value of 0.009 on a one-way ANOVA). Both isolates, FoSt01 and FoSt02, showed to be moderately pathogenic on tubers, but the first was more virulent than the later.
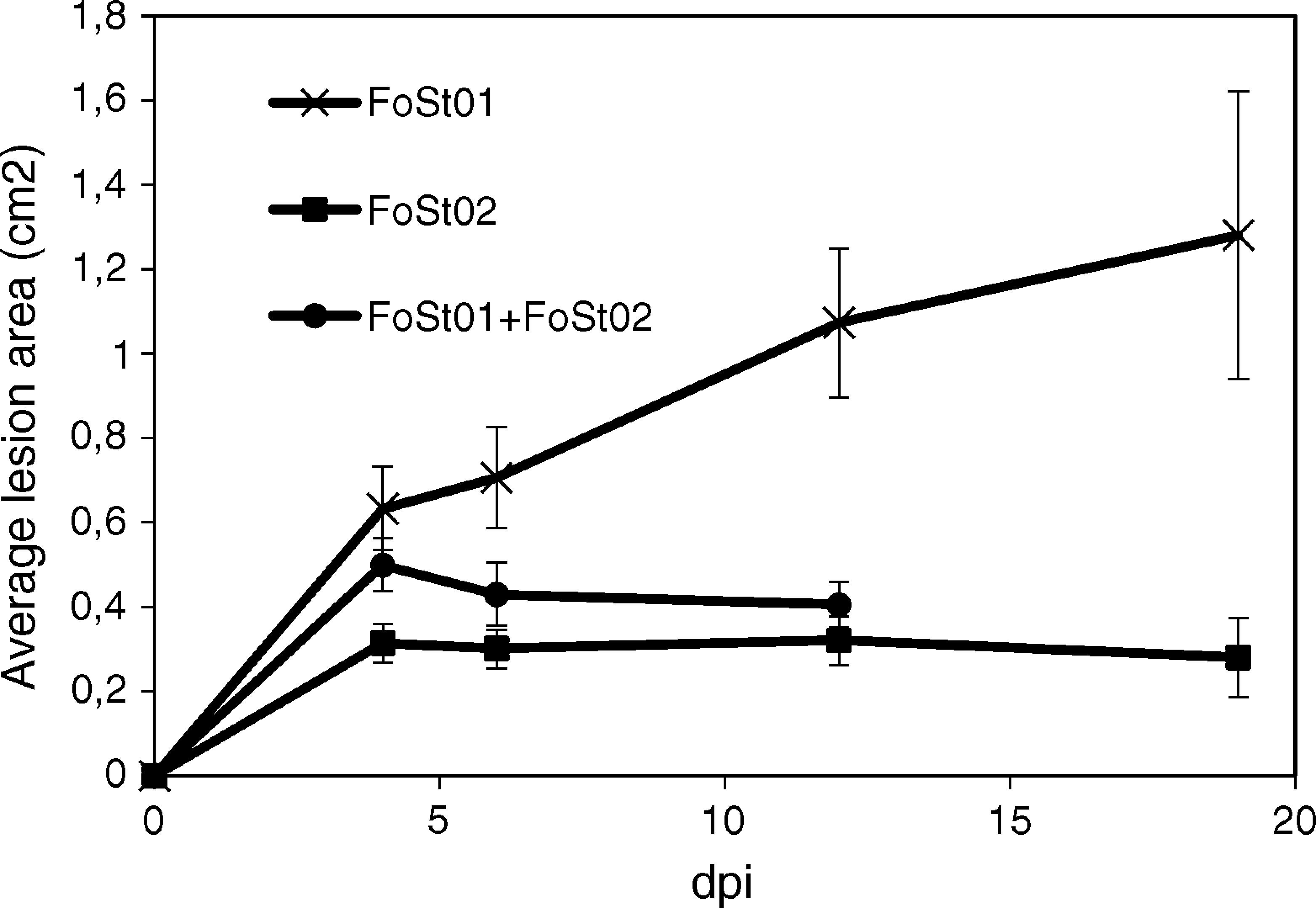
Trials on tuber slices were performed to observe the disease progression over time (Fig. 4). Co-inoculation of the two isolates was done to evaluate the effect of their interaction on virulent. A one-way ANOVA for each treatment over the different times of measurement indicated that there were no significant differences in the mean area of the lesions after 4 dpi (p>0.05), except for the treatment with FoSt01 alone, in which significant differences were observed. Comparisons between treatments were performed for only the 4 dpi due to the previous results. The mean lesion area was larger for FoSt01, followed by the co-inoculation treatment, and smallest for FoSt02 (p=0.007). Control tubers showed no measurable lesion development.
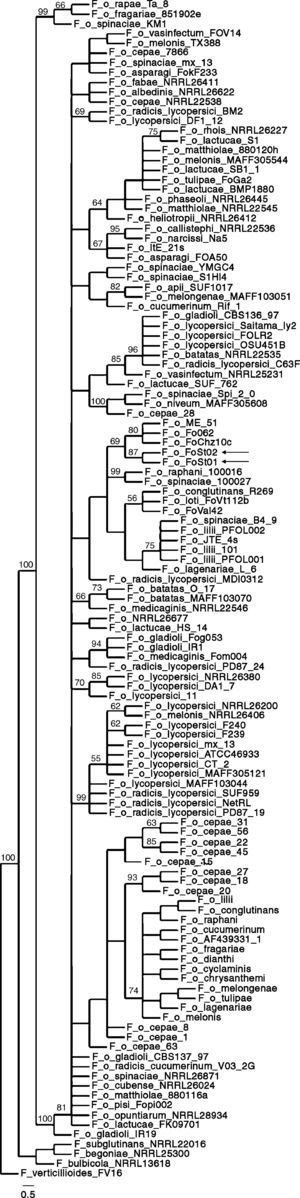
Phylogenetic analysisAs a first approach to a molecular characterization, ITS and IGS sequences of the ribosomal DNA were obtained. Both of them were identical between isolates. Similarity searches were done on the GenBank database, and the hits were used for the construction of maximum parsimony phylogenies. ITS sequences were consistent with both isolates being F. oxysporum but do not allow association to a forma specialis. The partial IGS sequences show a higher degree of polymorphism between formae speciales, and have therefore been used in association with other molecular markers to differentiate among them. IGS sequences showing a significant degree of similarity as determined by E-value on BLAST searches, were retrieved from GenBank, and were used to build a maximum parsimony phylogeny. The obtained consensus tree (Fig. 5) has 929 steps, 0.811 RI and 0.5705 CI. It proposes strains Fo062, FoChz10c and ME-51, retrieved from GenBank, as the most closely related to the two isolates in this study. Isolate Fo062 is pathogenic on red clover (Trifolium pratense), FoChz10c is a non-pathogenic isolate on birdsfoot trefoil (Lotus corniculatus)40 and ME-51 is a non-pathogenic isolate from Solanum lycopersicum.18
Red clover pathogenicity testsFor each inoculation or co-inoculation trial of both isolates on red clover (Trifolium pretense), ten replicates and two chronological repetitions were carried out. Both FoSt01 and FoSt02, alone and in co-inoculation, induced severe wilt in the two-week observation period, as compared to the healthy plants inoculated with sterile water. A generalized wilt was observed in the inoculated plants and when harvested, most of them showed root rot with vascular tissues involved. Both strains were successfully recovered from the diseased roots.
DiscussionAlthough F. oxysporum is not reported as one of the main causal agents of dry rot in most studied locations, its finding in this study does not come as a surprise, taking into account that it is indeed predominant in some countries outside the northern hemisphere.8,14,28,31 To the best of our knowledge, there are no previous reports on the incidence of dry rot on tubers in Colombia caused by F. oxysporum.
The recovery of two different, but closely related isolates, on a single diseased tuber is also not surprising, as it has been reported that a few mutations can lead to significant morphological differences between closely related isolates.20 On the other hand, the observation of differences in virulence remains to be explained. Intermediate lesion areas in co-inoculations might be explained by antagonism, probably competition for the same space and resources.1,38 Even if it remains to be explained why only for isolate FoSt02 disease progression stops after 4 days, this observation support the argument that although at the sequence level we could not be able to distinguish them, there are differences in virulence that should be explore at the molecular level.
Virulence of the two isolates is probably best described as moderate when compared to lesion areas reported for different isolates.7,8,14,31 However, one must keep in mind that the experimental conditions used in this study (relative humidity, incubation temperature and length of incubation period) might not be the optimal for the development of the disease by these isolates (assumed to be near 10°C, 95% RH and 6 weeks, respectively).7,30,31 Indeed, optimal disease progression conditions have been reported to vary significantly among strains.7 Thus, the isolates in this study could produce bigger lesions in tubers under storage for prolonged periods.
As mentioned, partial IGS sequences are far from being perfect molecular markers between formae specials, and identification of clonal lineages requires multilocus sequence typing (MLST) as well as vegetative compatibility group trials (VCG). MLST generally includes the complete intergenic spacer (IGS of rDNA [about 3,100bp]), an intron spanning region of the translation elongation factor 1 alfa (EF-1α), the mitochondrial small subunit rDNA (mtSSU), and mating type locus.13,21,24,40 Thus, the partial IGS sequences obtained in this study cannot lead us to conclude with complete certainty as to the forma specialis of the isolates. Moreover, many formae speciales being polyphyletic, mere association in a clade with similar partial IGS sequences cannot be directly interpreted as evidence for placing them in that forma specialis to the exclusion of others.21
All this said, the tree topologies observed in the reconstructed phylogenies conserved very similar topologies to trees built with MLST.13,24,40 In previous studies in which the same pair of primers were used, clonal lineages frequently showed 100% sequence identity.13 In addition, and probably more importantly, the closest relatives according to the maximum parsimony phylogeny (Fo062 and FoChz10) are also grouped together by MLST.40 Consequently, although the IGS sequences do not allow the assignment of the two isolates to a forma specialis, it can be safely stated that at least they are very close relatives to the above-mentioned isolates.
These are either pathogenic or non-pathogenic isolates of forage and tomato crops. This observation, although puzzling at first, has probably crop rotation as an underlying explanation. It has been reported that pathogenic and non-pathogenic isolates from cereal and forage crops in Canada (most notably one pathogenic isolate from Trifolium pretense) can also become pathogenic to potato.31 As cereals and forages are regularly rotated in a three-year cycle with potato, the strains probably gradually developed the ability to colonize this later host as a means of persistence.25 It has also been mentioned that for F. oxysporum, the major question remains to be whether non-pathogenic forms can evolve to become pathogenic and what would be the mechanisms underlying this transition.25
If that were the case, for the isolates obtained in this study, the question remains open as to whether the isolates have become regular pathogens of potato tubers or if forage crops continue to be the major reservoir. However, it is important to mention that, since no IGS sequences from F.o. tuberosi are available, it cannot be discarded that the two isolates belong to this forma specialis. Also, although the common source and identical ITS and IGS sequences support the hypothesis that the two isolates are most probably very closely related, VCG studies would be more conclusive.
All this comes to show that the two isolates obtained from the diseased tuber might be an evidence of transfer from forage species, which could be gradually developing the capability to infect tubers. This is supported by the pathogenicity of both isolates on red clover. As previously mentioned, the polyphyly of several formae speciales can be explained by several separate events of colonizing a new host.25 Thus there is a need to further characterize not only these strains, but the pathogen population causing Fusarium dry rot on potatoes in Colombia. This way, it could be established if the pathogen populations are currently steady or if the emergence of new pathogenic isolates could lead to a higher incidence of the disease.
Conflict of interestThe authors report no conflict of interest.