Sporothrix schenckii, an ascomycetous dimorphic organism that for over a century was recognized as the sole agent of sporotrichosis, a subcutaneous mycosis with a worldwide distribution. However, it has been proposed, based on physiologic and molecular aspects, that S. schenckii is a complex of distinct species: Sporothrix brasiliensis, Sporothrix mexicana, Sporothrix globosa, S. schenckii sensu strictu, Sporothrix luriei, and Sporothrix albicans (formerly Sporothrix pallida). Human disease has a broad range of clinical manifestations and can be classified into fixed cutaneous, lymphocutaneous, disseminated cutaneous, and extracutaneous sporotrichosis. The gold standard for the diagnosis of sporotrichosis is the culture; however, serologic, histopathologic and molecular approaches have been recently adopted for the diagnosis of this mycosis. Few molecular methods have been applied to the diagnosis of sporotrichosis to detect S. schenckii DNA from clinical specimens, and to identify Sporothrix spp. in culture. Until now, Sporothrix is the unique clinically relevant dimorphic fungus without an elucidated genome sequence, thus limiting molecular knowledge about the cryptic species of this complex, and the sexual form of all S. schenckii complex species. In this review we shall focus on the current diagnosis of the sporotrichosis, and discuss the current molecular tools applied to the diagnosis and identification of the Sporothrix complex species.
This manuscript is part of the series of works presented at the “V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi” (Oaxaca, Mexico, 2012).
Sporothrix schenckii, un organismo ascomiceto dimorfo que durante más de un siglo fue reconocido como el único agente de esporotricosis, una micosis subcutánea con una amplia distribución mundial. Sin embargo, se ha propuesto, con base en los aspectos fisiológicos y moleculares, que S. schenckii es un complejo de especies distintas: Sporothrix brasiliensis, Sporothrix mexicana, Sporothrix globosa, S. schenckii sensu stricto, Sporothrix luriei y Sporothrix albicans (antes Sporothrix pallida). La enfermedad humana tiene una gama de manifestaciones clínicas y puede clasificar e cutánea fija, linfocutánea, cutánea diseminad y esporotricosis extracutánea. El estándar de oro para el diagnóstico de esporotricosis es el cultivo del patógeno, sin embargo, los métodos serológicos, histopatológicos y, recientemente, los moleculares se han usado para el diagnóstico de esta micosis Pocos métodos moleculares han sido aplicados para el diagnóstico de la esporotricosis para detectar ADN de S. schenckii a partir de muestras clínicas y para identificar Sporothrix spp. en cultivo. Hasta ahora, Sporothrix es el único hongo dimorfo clínicamente relevante sin una secuencia genómica elucidada, limitando el conocimiento molecular de las especies crípticas de este complejo y la forma sexual de todas las especies del complejo S. schenckii. Esta revisión se centrará en el diagnóstico actual de la esporotricosis con énfasis en las herramientas moleculares vigentes aplicadas tanto al diagnóstico como en la identificación de las especies del complejo Sporothrix. Este artículo científico es parte de la serie de trabajos presentados en el «V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi» (Oaxaca, México, 2012).
Sporothrix schenckii is an ascomycetous dimorphic organism (Ascomycota, Pyrenomycetes, Ophiostomatales, Ophiostomataceae) that is found in substrates like living and decayed vegetation, animal excreta, and soil.5,19 This fungus is phenotypically characterized by the ability to produce sessile dematiaceous conidia along with hyaline sympodial conidia in its filamentous form, and cigar-shaped yeast-like cells in parasitism or when cultured at 35–37°C on specific culture media.5,37
For over a century, this species was recognized as the sole agent of sporotrichosis, a subcutaneous mycosis with a worldwide distribution.34 However, it has been proposed, based on physiologic and molecular aspects that S. schenckii is a complex of four distinct species: Sporothrix brasiliensis, Sporothrix mexicana, Sporothrix globosa, and S. schenckii sensu strictu.37 More recently, S. schenckii var. luriei was considered to be another species belonging to the S. schenckii complex, and was therefore named Sporothrix luriei.38 Additionally, in other phylogenetic analysis from Sporothrix albicans, Sporothrix pallida, and Sporothrix nivea revealed a significant similarity among them. Therefore, it has been proposed that all these three non-pathogenic species closely related to S. schenckii were called S. pallida.12,54 So, S. schenckii sensu strictu, S. brasiliensis, S. globosa, S. mexicana, and S. luriei are now recognized as agents of sporotrichosis.
Sporotrichosis affects humans and other mammals, such as dogs, cats, rats, armadillos, and horses.5,34 Human disease has a broad range of clinical manifestations and can be classified into fixed cutaneous, lymphocutaneous, disseminated cutaneous, and extracutaneous sporotrichosis.5 Another clinical type of this infection is the primary pulmonary sporotrichosis, which results from inhalation of conidia.49 Lymphocutaneous sporotrichosis is the most common manifestation and is characterized by lesions in the site of injury and along the lymph vessels path. These lesions can be papular, nodular, gummy (ulcerated or not), and/or vegetative plaque. In fixed cutaneous sporotrichosis, the lesion is restricted to the injury site, with no lymphatic involvement. Disseminated cutaneous form consists in several lesions in the body after hematogenous dissemination or multiple inoculation of the fungus into the skin. In extracutaneous disease, several organs or tissues can be affected, such as bones, lungs, eyes, and central nervous system.62 Along with these manifestations, other clinical aspects have been recently reported such as erythema nodosum,21 erythema multiforme,20 and Sweet's syndrome.16
Definitive diagnosis of sporotrichosis is based on fungal detection in culture.62 Microscopic methods for yeast detection in clinical samples are of low sensitivity5 and asteroid body observation varies between different works.17,50 Cultivation of clinical specimens in mycological media such as Sabouraud dextrose agar or mycobiotic agar yields white filamentous colonies that become brown to black after a few days. Subculturing these colonies in brain heart infusion at 35–37°C results in white to creamy yeast-like colonies.5,62S. schenckii identification is based on its macro- and micromorphologies of the mycelial and yeast forms. These characteristics, however, do not differentiate the new described species on the Sporothrix complex. In order to physiologically differentiate the species within this complex, other tests such as carbohydrate assimilation (especially sucrose and raffinose), growth rates at 30°C and 37°C as well as production of dematiaceous conidia are necessary.37,38 However, discrepancies between physiological and molecular methods of identification have been described.47
Several techniques have been described for the establishment of immunological diagnosis of sporotrichosis based on antibody detection in sera from infected patients. Precipitation and agglutination methods were first employed in the diagnosis1 and more recently immunoenzymatic assays are used in sporotrichosis serodiagnosis.2,8 However, there is no immunological method described up to now that permits differentiation of the species of the Sporothrix complex. Recent publications showed that the species of the S. schenckii complex differ in antifungal susceptibility and virulence.4,22,40 These publications have suggested that even lesion mechanisms could be related to these different species. Therefore, molecular methods which allow rapid identification and differentiation of these close-related fungi are very necessary to the mycology laboratory.
Molecular methods for sporotrichosisPCR diagnosis based on the amplification of the fungal gene sequences is a powerful tool for identifying invasive mycoses. The first PCR for identification of S. schenckii sensu lato was described by Kano et al.29 Specific oligonucleotide primers based on the chitin synthase 1 gene were designed, and PCR was able to detect a 10pg genomic DNA fragment of S. schenckii. After, this methodology was employed for the molecular diagnosis of sporotrichosis.28 A nested PCR assay for the detection of S. schenckii was evaluated in clinical samples using the 18S rRNA gene sequence as the target. The nested PCR could detect S. schenckii DNA in tissue samples from infected animals or from clinical specimens from patients with sporotrichosis confirmed by culture or hystochemical staining. The test showed high sensitivity and specificity, indicating that the assay could provide rapid diagnosis with sufficient accuracy to be clinically useful for patients with sporotrichosis.25 More recently, the same assay was used to detect S. schenckii DNA from 38 strains (including all 24 mitochondrial DNA types) collected from different areas of the world, and of tissues of experimentally infected mice, skin biopsies of nine patients with sporotrichosis, two strains of Ceratocystis minor, and 10 different species of other pathogenic fungi.61 The authors demonstrated that nested PCR could correctly identify all S. schenckii strains and clinical samples, corroborating the data obtained by Hu et al.25 that the nested PCR assay is highly sensitive and specific and is a rapid method for diagnosis of sporotrichosis under contamination free conditions.61 Other studies also reported PCR methods applied in the diagnosis of sporotrichosis. Mendoza et al.41 showed an evolution in the previously described nested PCR.25 However, this study demonstrated that this molecular methodology presented lower efficacy than conventional diagnostic methods, such as the gold standard culture. Liu et al.33 reported a PCR in biopsy tissue using the primer S2-R2 that showed positive bands in twenty-five out of the thirty cases (83.3%). These primers were also successfully used for the diagnosis of feline sporotrichosis, with no cross-reactions with normal cat skin cells or other dimorphic fungi.30 These studies did not permit classification at species level of the Sporothrix complex.
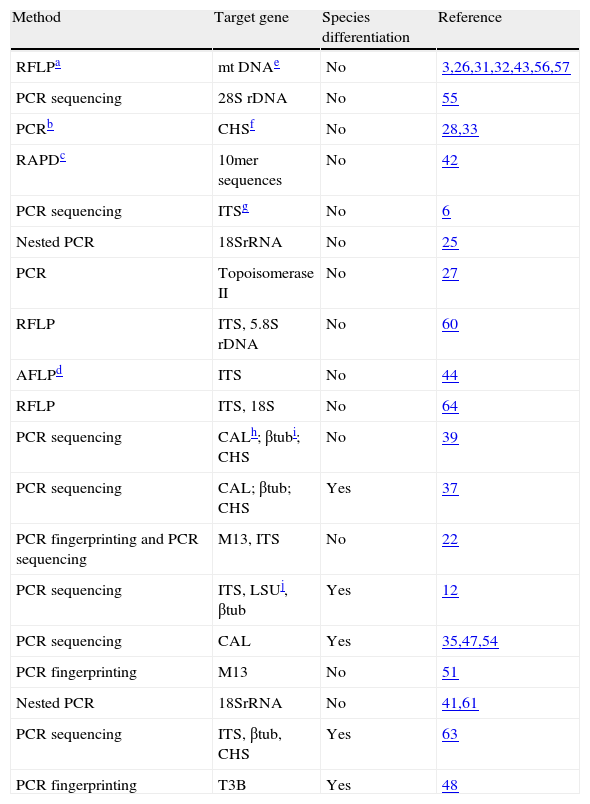
Molecular identification of Sporothrix speciesIn the last few years, the development of DNA-based methods to identify fungal isolates has led to a decrease in the time-consuming step of morphological identification, while maintaining or improving the specificity, accuracy and sensitivity. Until now, few molecular methods have been applied in the detection of S. schenckii DNA from clinical specimens, and in the identification of Sporothrix spp. in culture (Table 1). Diagnosis using PCR is based on the amplification of fungal gene sequences and is a powerful tool for identifying mycoses. One of the pioneering DNA-based methodologies used for the diagnosis of fungal infections was reported by Sandhu et al.55 who developed 21 specific nucleic acid probes targeting the large subunit of the rRNA gene from several fungi, including S. schenckii. The results show a high level of specificity.55 But preview studies56,57 using mitochondrial DNA analysis have reported heterogeneity profiles in S. schenckii strains. In fact, several molecular taxonomic studies using different methodologies, such as restriction fragment length polymorphism (RFLP) from different gene targets, random amplified polymorphic DNA (RAPD), DNA sequencing of internal transcriber spacer (ITS) regions of the ribosomal RNA gene (rRNA), PCR targeting the DNA topoisomerase II gene, amplified fragment length polymorphism (AFLP), and M13 PCR fingerprinting have demonstrated that S. schenckii isolates present different genetic characteristics, suggesting that they do not belong to the same species.3,26,27,32,39,43,44,56,57,60,64
Molecular studies on the Sporothrix schenckii complex.
| Method | Target gene | Species differentiation | Reference |
| RFLPa | mt DNAe | No | 3,26,31,32,43,56,57 |
| PCR sequencing | 28S rDNA | No | 55 |
| PCRb | CHSf | No | 28,33 |
| RAPDc | 10mer sequences | No | 42 |
| PCR sequencing | ITSg | No | 6 |
| Nested PCR | 18SrRNA | No | 25 |
| PCR | Topoisomerase II | No | 27 |
| RFLP | ITS, 5.8S rDNA | No | 60 |
| AFLPd | ITS | No | 44 |
| RFLP | ITS, 18S | No | 64 |
| PCR sequencing | CALh; βtubi; CHS | No | 39 |
| PCR sequencing | CAL; βtub; CHS | Yes | 37 |
| PCR fingerprinting and PCR sequencing | M13, ITS | No | 22 |
| PCR sequencing | ITS, LSUj, βtub | Yes | 12 |
| PCR sequencing | CAL | Yes | 35,47,54 |
| PCR fingerprinting | M13 | No | 51 |
| Nested PCR | 18SrRNA | No | 41,61 |
| PCR sequencing | ITS, βtub, CHS | Yes | 63 |
| PCR fingerprinting | T3B | Yes | 48 |
Hence, Marimon et al.37 supported these findings describing through a combination of phenotypic and genetic features four S. schenckii-related species: S. brasiliensis, S. globosa, S. luriei, and S. mexicana. S. globosa has a worldwide distribution,11,35,46 whereas S. mexicana is restricted to Mexico and S. brasiliensis to Brazil.37 However, a case of human sporotrichosis due to S. mexicana was reported in 2011 by Dias et al. in Portugal.13 The molecular tool proposed to differentiate these closely related species was partial calmodulin (CAL) gene sequencing.37 Therefore, an identification key for the S. schenckii complex that includes morphologic analysis of conidia, auxonogram, genotyping via PCR amplification, and sequencing of the CAL gene. Based on this last analysis, Romeo et al.,54 who were studying the molecular phylogeny and epidemiology of Sporothrix species isolated in Italy, demonstrated that 26 environmental strains co-clustered with S. albicans, and two clinical isolates grouped with S. schenckii sensu stricto.
Additionally, phylogenetic analysis based on the rDNA and the β-tubulin regions from S. albicans, S. pallida and S. nivea revealed a significant similarity among them. Therefore, it has been proposed that all the three species were called S. pallida. Moreover, other three environmental Sporothrix species were described using this molecular approach: Sporothrix stylites, Sporothrix humicola and Sporothrix lignivora. The two first species differ from S. schenckii by production of only hyaline conidia and consequent no darkening of colonies with age. S. lignivora has distinctive conidia that do not match in size and shape with other Sporothrix or Ophiostoma species. Isolates classified as S. humicola in this study were referred before as environmental isolates of S. schenckii. In their study, the authors conclude that β-tubulin sequence analysis is strongly recommended in taxonomic studies from Sporothrix species isolated from the environment.12 In fact, β-tubulin analysis, together with ITS sequencing enable the further description of other two environmental Sporothrix species: Sporothrix brunneoviolacea and Sporothrix dimorphospora.36
After the description of Sporothrix complex, one important question is the search for rapid methods for species identification and typing. Oliveira et al.48 reported a PCR fingerprinting using the universal primer T3B to distinguish among species of the Sporothrix complex. The T3B fingerprinting generated clearly distinct banding patterns, allowing the correct identification of all 35 clinical isolates at the species level, which was confirmed by partial CAL gene sequence analyses. Overall, there was a 100% agreement between the species identification using both genotypic methodologies. These profiles were also able to accurately distinguish the species misidentified by phenotypic analysis. The proposed identification technique is simple, reliable, more rapid, less expensive, and requires less technical expertise than sequencing. The computer scanned PCR profiles generated can form the basis of a computer database, which can be used for future identifications of atypical or unidentifiable Sporothrix isolates. This methodology is supposed to be an ideal routine identification system for clinical mycology laboratories, particularly those with limited facilities or technical expertise. There are only a few molecular tools for Sporothrix typing,31,51 and the development of new typing methods are necessary.
Other molecular aspects of SporothrixThe sexual form of all S. schenckii complex species is unknown nowadays; however, there are strong molecular evidences that this fungus undergoes recombination in nature.42 Molecular analyses from 18S region of ribosomal DNA have shown indirect evidence that the sexual form of S. schenckii was Ophiostoma stenoceras.7 On the other hand, morphological and physiological studies show consistent differences between these two species.14,45 These observations lead to consider as different species the O. stenoceras anamorph and S. schenckii. Meanwhile, other molecular studies6,12,24 reinforce that the S. schenckii teleomorph belongs to the genus Ophiostoma, but it is different from O. stenoceras.
Molecular analysis of S. schenckii can also be linked with fungal virulence. A study about the ribosomal conserved region (i.e., ITS) and the nontranscribed spacer (NTS) demonstrated that a S. schenckii sensu strictu strain from disseminated cutaneous sporotrichosis presents a 10bp deletion in the ribosomal NTS region, when compared to isolates controls obtained from patients with fixed cutaneous sporotrichosis.63 Nucleotide polymorphisms are also able to separate environmental and clinical S. schenckii strains. Two single-base transitions in the D1–D2 domain of rDNA differentiate strains from these groups.10
Rapid amplification of cDNA ends technique was recently applied to characterize the gene sequence of a physiology-related protein in S. schenckii. A progesterone adiponectinQ receptor was found to be responsible for an inhibition of, especially, the yeast form of the fungus.18 However, it is necessary a large investigation about this inhibition, since there are several cases of sporotrichosis in woman, pregnant or not,5,9,15 particularly in the endemic area of sporotrichosis where S. brasiliensis predominates.47
S. schenckii is the only clinically relevant dimorphic fungus without an elucidated genome sequence, thus limiting molecular knowledge about the cryptic species of this complex.53 However, some genes have been recently described, such as the α-subunit of the endoplasmic reticulum glucosidase II,52 the α1,2-mannosyltransferase,23 a cytosolic phospholipase A2,58 and a calcium/calmodulin kinase gene.59 These sequences are potential targets for the development of new identification and typing methods to be applied within the Sporothrix complex species.
ConclusionNevertheless, nucleotide probes, specific for the Sporothrix species complex, and DNA amplification procedures, such as PCR, allow more rapid and precise diagnosis, which can lead to earlier treatment. The gold standard in diagnosis continues to be using the culture with the correlation of phenotypic characteristics and molecular data.
Conflict of interestThe authors declare no conflict of interest.
Financial support was partially provided by FAPERJ (grant proc. E-26/110.619/2012). R. M. Z-O is supported in part by CNPq350338/2000-0 and FAPERJE-26/103.157/2011. M. M. E. O was supported by a grant from CAPES2445/11-5. The authors thank Dr. Maria Lucia Taylor for her editorial assistance in the Spanish language.
These authors contributed equally to this work.