Late blight, caused by Phytophthora infestans, is one of the most devastating diseases found in potato and tomato crops worldwide. In Colombia it also attacks other important crops: cape gooseberry and tree tomato. The knowledge of the pathogen population is determinant to effectively design control strategies.
AimsTo determine the physiological and molecular characteristics of a set of Colombian P. infestans isolates.
MethodsStrains isolated from Cundinamarca and Boyacá were examined for the level of resistance to mefenoxam and cymoxanil. Virulence was tested for all strains and crosses between A1 mating type, from different hosts, and the Colombian A2 mating type were tested for the production and viability of oospores in different substrates. Additionally, the molecular diversity of the avirulence gene Avr3a, the β-tubulin gene, and two single copy genes showing RxLR motif, was assessed.
ResultsWe found all levels of mefenoxam sensitivity, with 48% of the strains resistant. A high diversity of races was detected and the population was genetically clonal. Colombian strains had the possibility of sexual reproduction.
ConclusionsThese results will help in optimizing the use of fungicides and deployment of resistance as control strategies and will contribute to broader studies on diversity of this pathogen.
El tizón tardío, causado por Phytophthora infestans, es una enfermedad devastadora de la papa y el tomate a nivel mundial, y en Colombia también ataca otros cultivos como la uchuva y el tomate de árbol. El conocimiento de la población del patógeno es determinante para el diseño efectivo de estrategias de control.
ObjetivosDeterminar las características fisiológicas y moleculares de aislamientos colombianos de P. infestans.
MétodosEl nivel de resistencia al mefenoxam y al cimoxamil fue evaluado en aislamientos de Cundinamarca y Boyacá. Se estimó su virulencia y se determinó la producción y viabilidad de oosporas en diferentes sustratos con cruces entre aislamientos A1 y el aislamiento colombiano A2. Además, se determinó la diversidad molecular en el gen de avirulencia Avr3a, el gen de la β-tubulina y otros dos genes de copia única con motivo RXLR.
ResultadosLos aislamientos colombianos tuvieron la posibilidad de reproducirse sexualmente. Encontramos todos los niveles de sensibilidad al mefenoxam, con el 48% de los aislamientos resistentes. Se detectó una diversidad de razas y a nivel genético la población fue clonal.
ConclusionesEstos resultados ayudarán a optimizar el uso de fungicidas y reducir la resistencia como estrategias de control, además de contribuir al conocimiento de la diversidad de este patógeno.
The knowledge of pathogen population structure and changes that these populations can experience over time is crucial for developing durable crop disease management strategies.14 Due to variability of conditions in each country, region and even within each field, detailed local population studies must be carried out. This is especially true when disease control measures at a certain time and location are not effective or when they represent an economical or environmental burden.
Late blight is one of the most devastating plant diseases demanding a high chemical input for disease control worldwide.11 This disease, caused by the oomycete Phytophtora infestans, affects a wide range of solanaceous plants. In Colombia, cultivated solanaceous plants such as Solanum tuberosum (potato), Solanum phureja (yellow potato), Solanum lycopersicum (tomato), Solanum betaceum (tree tomato), Solanum quitoense (lulo) and Physalis peruviana (cape gooseberry) are P. infestans hosts.34 Optimal conditions for both pathogen and disease development, of particular importance temperature and humidity, are found everywhere potatoes are cultivated in Colombia. In addition, main potato cultivars grown in the regions Diacol Capiro, Parda Pastusa, Tuquerreña and criolla are highly susceptible to the disease.12 Recently, severe epidemics have been reported in lulo and tree tomato crops.37
Chemical applications are the preferred strategy to control late blight,5 and mixtures of systemic and protective fungicides are the most widely used.27 Two molecules with systemic activity usually incorporated in fungicide mixtures are mefenoxam, the most active enantiomer contained in the fungicide Ridomil Gold™, which inhibits ribosomal RNA synthesis, and cymoxanil whose mode of action is still unknown.27 Mefenoxam (also known as metalaxyl-M) is comprised almost solely of the R enantiomer of the molecule, while metalaxyl is a mixture composed of approximately equal proportions of the R and S enantiomers. Metalaxyl resistance has evolved rapidly ever since its introduction in 1980, with resistance being detected first in Ireland and The Netherlands.6 Since this first report, several studies worldwide were conducted to explain the genetic resistance to metalaxyl,8,20,21 the presence of resistant strains in the populations and the effect of numerous chemicals that may aid in reducing the spread of the disease.15,22,29 Resistance values for this fungicide have risen up to 100mg/l in countries where this fungicide is commonly used.30 Although P. infestans has been thought to be sensitive to cymoxanil, Grünwald et al. showed that in Mexico some isolates exhibited resistance and a selection pressure directed to resistance was observed.18 The least sensitive strain reported was one isolate from Israel, which showed an EC90 value of 467mg/l. In other countries such as Mexico, The Netherlands and Ireland these values ranged between 64 and 152mg/l for the least sensitive strains.31
In Colombia pathogen populations are dominated by the EC-1 clonal lineage, and a couple of isolates were determined to be variants of this clonal lineage (e.g. EC1.1) and the clonal lineage, CO1. Mating type A1 and mitochondrial haplotype IIa were the most commonly found in a previous study and isolates can be recovered from a wide diversity of hosts.35 A Colombian isolate from cape gooseberry was recently characterized as US-8, mitochondrial haplotype Ia and mating type A2, indicating the possibility of sexual reproduction in Colombia.30,35
As a consequence, we established the potential of producing viable sexual progeny by making crosses between isolates classified as A1 mating type from different hosts and the Colombian A2 mating type from cape gooseberry. In addition, to further characterize a sample of Colombian P. infestans isolates gathered from the Central Andean region, we established their fungicide baseline sensitivity to two molecules (mefenoxam and cymoxanil) and we characterized their molecular diversity in the avirulence gene Avr3a,1 the β-tubulin gene and two single copy potential RxLR effector genes.36 Pathogen effectors are thought to co-evolve in an arms race with corresponding host resistance genes. Because effectors are important determinants in virulence and pathogenicity, we investigated selected gene sequences looking for polymorphisms, which may help to explain host virulence, indicate co-evolution or be used as potential markers for clonally reproducing populations of this pathogen.
Materials and methodsIsolate collection and maintenanceTwenty-five isolates were obtained from the P. infestans culture collection maintained in the Laboratory of Mycology and Plant Pathology at Universidad de los Andes (LAMFU), 21 from Colombia and two from Venezuela. Isolates mating type, mitochondrial haplotypes and genotypes for several microsatellites have been previously reported.35 Colombian isolates were chosen with the aim of selecting the most diverse ones in the central states of Boyacá and Cundinamarca, based on the characteristics mentioned above (Table 1). Isolates US940480 and US940494 were provided by William E. Fry (Cornell University) and were used as reference strains. Isolates were maintained in rye A agar at 14°C.12
Phytophthora infestans strains used in this study.
| Host of origin | Isolate no.a | Collected site | Clonal lineageb | Mating typeb | Mitochondrial haplotypeb | Mefenoxam sensitivityc | Race |
| Solanum tuberosum | 1810 | Boyacáe | EC-1 | A1 | IIa | R | 1.3.4.5.6.7.10 |
| S. tuberosum | 2400 | Granadae | EC-1 | A1 | IIa | R | 1 |
| S. tuberosum | 2401 | Coguae | EC-1 | A1 | IIa | I | 2.3.4.5.6.7.8.1.11 |
| S. tuberosum | 2414 | Coguae | EC-1 | A1 | IIa | S | 10 |
| S. tuberosum | 2421 | Coguae | EC-1 | A1 | IIa | S | 2.3.4.8 |
| S. tuberosum | 2435 | Granadae | EC-1 | A1 | IIa | S | 1.7 |
| S. tuberosum | 2404d | Coguae | Nd | A1 | IIa | Nd | Nd |
| S. tuberosum | US940480 | Reference strain | US-8 | A2 | Ia | S | Nd |
| S. phureja | 2473 | Coguae | EC-1 | A1 | IIa | S | 2.4.7.8.10 |
| S. phureja | 2475 | Coguae | EC-1 | A1 | IIa | I | 2.3.5.7.8 |
| S. phureja | 2506 | Granadae | EC-1 | A1 | IIa | S | Nd |
| S. phureja | 2518 | Granadae | EC-1 | A1 | IIa | S | 2.3 |
| S. phureja | 2526 | Granadae | EC-1 | A1 | IIa | S | 2.3.5.6.8.10.11 |
| S. phureja | 2524d | Granadae | Nd | A1 | Nd | Nd | Nd |
| S. phureja | Z3-2 | Zipacóne | Nd | A1 | Nd | S | Nd |
| S. phureja | 6 | Guascae | Nd | A1 | Nd | I | 1.5.7.11. |
| S. phureja | 10 | Guascae | Nd | A1 | Nd | S | 1.3.5.6.7.10.11. |
| S. phureja | 21 | Guascae | Nd | A1 | Nd | I | 7.8.10. |
| S. phureja | 100 | Guascae | Nd | A1 | Nd | R | 5.7.10.11. |
| S. lycopersicum | 2527d | Tabioe | Nd | A1 | Nd | Nd | Nd |
| S. lycopersicum | US940494 d | Reference strain | US-12 | A1 | IIb | Nd | Nd |
| Physalis | |||||||
| peruviana | 2557 | San Franciscoe | CO-1 | A1 | IIa | S | 5.7.10.11. |
| P. peruviana | 2562 | San Franciscoe | EC-1 | A1 | IIa | R | 1.5.6.7.8.10. |
| P. peruviana | 2768 (4084)f | San Franciscoe | US-8 | A2 | Ia | S | 2.3.5.6. |
| Solanum lycopersicum | 2530 | Tibacuye | EC-1 | A1 | IIa | R | Nd |
| S. tuberosum | SR1d | Santa Rosa, Mérida, Venezuela | Nd | A1 | Ia | Nd | Nd |
| S. tuberosum | TCH2d | Las Porqueras, Táchira, Venezuela | Nd | A1 | Ia | Nd | Nd |
Identification number of the P. infestans isolate in the Phytophthora collection at La Universidad de los Andes, Colombia.
Distinctive characteristics such as Genotype, Mating type and mtDNA haplotype. Previously determined in Vargas et al.35
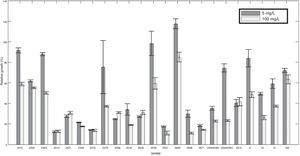
Mefenoxam sensitivity determined in vitro using agar media amended with 0.5 and 100mg of mefenoxan technical grade 95% per liter. Sensitive isolates (S)≤40% relative to mycelia growth (RMG) at 5 and 100mg/l. Intermediate isolates (I)≥40% RMG at 5mg/l and <40% RMG at 100mg/l. Resistant isolates (R)≥40% RMG at 5 and 100mg/l of mefenoxam.
The sensitivity of P. infestans isolates to the phenylamide fungicide mefenoxam as technical grade (90%) was evaluated using the radial growth assay method.17 Each isolate was transferred to rye B agar medium amended with mefenoxam at 0.5 and 100mg/l. One plug of 5mm in diameter was placed on the center of each plate. Mycelial radial growth in each plate was recorded 7 days after inoculation. The relative growth of each isolate growing in fungicide-amended media was obtained by subtracting the plug diameter from the diameter of the colony (after 7 days), and then dividing the corrected radial growth of the isolate growing in the fungicide-amended media by the radial growth of the same isolate growing in media without fungicide. Colony diameters were determined as the average diameter between two measured diameters. One of those diameters was determined by measuring one random diameter, and then a second diameter that was perpendicular to the first. Isolates were scored as resistant when radial growth at 5 and 100mg/l was more than 40% of the growth without mefenoxam, as intermediate when the radial growth was more than 40% at 5mg/l but less than 40% with 100mg/l and as susceptible when radial growth was less than 40% in both 5 and 100mg/l.10 These concentrations and ranges were selected because of the good resolution they provide to distinguish differences between the different levels of metalaxyl sensitivity in previous studies.21–23 Three replicates were performed per concentration for each isolate, and the experiment was repeated twice. In addition, sensitivity to commercial products that contain mancozeb besides the systemic molecule Ridomil Gold (ai=Mefenoxam) (Syngenta, NC, USA) and Curzate (ai=Cymoxanil) (DuPont, DE, USA) was also evaluated with similar assays, except that the concentrations tested were 0.8 and 1.2mg/l for both fungicides. The Effective Concentration for which 50% of the mycelial growth was inhibited (EC50) was calculated for each fungicide by Probit analysis using the computer software Statgraphics Centurion XVI (Statpoint Technologies, VA, USA).
Oospores assaysIn vitro oospore production and viability assay. Colombian A1 mating type isolates, A2 US940480 and Colombian A2 mating type isolate 2768 were grown on rye B agar. After seven days, a 0.5cm plug of each A1 and A2 mating type isolates was placed 2cm apart from each other on a rye B agar plate. Crosses were kept at 18°C in the dark and from day fourteen until day 30, when they were checked for oospores production using a light microscope. On day thirty, 1-cm2 was excised from the area where mycelium from both mating types was encountered. The agar square was placed on a microscope slide; one drop of distilled sterile water was deposited on top of it, before covering it with a cover slide. The oospores present all over the agar square were counted under light microscopy (40×). When counting, different depths were considered by using the fine adjustment knob given that some oospores were superficially exposed while others were embedded in the agar. The number of oospores produced by each cross performed was normalized using the natural logarithm (ln) transformation. An ANOVA test was conducted to determine whether the production of oospores was dependent on the A2 mating type isolate used (Colombian versus reference).
For the viability assay a modification of the Pittis and Shattock plasmolysis protocol was followed.28 After oospore counting, the 1cm2 agar square was placed inside a 1.5ml eppendorf tube and was ground with the help of a pestle, after addition of 1ml of 4M NaCl to induce plasmolysis of the viable oospores at room temperature. Three hours later, 20μl of this homogenized mixture were placed on a microscope slide and the percentage of plasmolyzed versus not-plasmolyzed oospores was determined. Three replicates were mounted per isolate and the whole experiment was repeated three times. Due to the fact that some P. infestans isolates can produce oospores in the absence of the opposite mating type, isolates used in this study were tested for self oospore production as a control, to ensure that the observed oospores were the result of the sexual reproduction of the tested isolates.32
Oospore production in a detached leaf assay. Sporangial suspensions were obtained for each of the A1 and the two A2 mating type isolates studied. For this, fifteen-day old P. infestans cultures grown on rye B agar were washed with 3ml of distilled sterile water and sporangia concentration adjusted to 2000 sporangia/ml using a hemocytometer. The sporangial suspension was subsequently placed at 4°C for two hours to allow zoospore release. Ten μl of each mating type combination were placed one cm apart from each other, at both sides of the main vein of the abaxial side of a S. phureja leaflet that was placed inside a moist chamber. Once the inoculated leaflets began to develop typical late blight symptoms, they were monitored for the presence or absence of oospores until three weeks after inoculation.
Oospore production in soil. Sporangial suspensions were obtained as mentioned above and the concentration was adjusted to 10,000 sporangia/ml. One mililiter of each isolate was added to sterilized soil placed inside a 50ml plastic sterile cup. Each A1 isolate was crossed with both US94080 and Colombian 2768 A2 isolates. After one month, the inoculated soil was sieved through different pore sizes, retained at the 10μm pore size sieve and was finally resuspended in 100ml of sterile distilled water. Suspension was vacuum filtered using Whatman #1 filter paper. Whatman paper with oospores was washed with 1ml of water and observed under light microscope to search for oospores. At the same time, the filtrate was concentrated by centrifuging it at 8000rpm and the pellet was resuspended in 1ml of sterile distilled water and observed under light microscopy to record oospore presence.
Virulence testsVirulence was determined using the detached leaf assay described before.29 Each isolate was challenged against an R-gene differential set of potato clones, each one carrying a single resistance gene (R1–R11), in order to determine virulence and race of P. infestans isolates.18 Four detached leaflets of each differential were placed abaxial side up in moist chambers and inoculated with 20μl of a sporangial suspension (aprox. 20,000sporangia/ml−1) of each isolate. The leaflets were incubated in growth chambers at 18°C, 85% relative humidity and 14h light. Seven days after inoculation, leaflets were evaluated for presence of sporulation and rated as compatible or incompatible.9
Molecular characterizationIn order to allow mycelia production, isolates were grown in pea broth for 15 days at 18°C. DNA extraction was performed as described by Goodwin et al.16 The β-tubulin gene, Avr3a and two single-copy RXLR genes, SC9 and SC1636 were amplified. All PCR reactions were carried out in a final volume of 25μl, with 1X PCR buffer, 0.5mM dNTPs, 2.5mM MgCl2, 0.2mM of each primer and 1U Taq DNA polymerase (Fermentas Life Sciences). PCR conditions for the β-tubulin gene started with a denaturation step of 2min at 94°C, followed by 35 cycles of 94°C for 30s, 60°C for 30s, 72°C for 1min and a final step of 10min at 72°C. For the amplification of the Avr3a gene we followed the protocol described in Cárdenas et al.4 with the modified primer set (M13Pex F: 5′-GTA AAA CGA CGG CCA GCC ATG CGT CTG GCA ATT ATG CT-3′ and M13Pex R: 5′-CAG GAA ACA GCT ATG ACC TGA AAA CTA ATA TCC AGT GA-3′).4 Amplification conditions included a denaturation step of 2min at 94° C followed by 15 cycles of 30s at 94°C, 30s at 55°C and 1min at 72°C. Twenty-five additional cycles of 30s at 94°C, 30s at 62°C and 1min at 72° C were performed. The final extension was carried out for 10min at 72°C. For SC9 and SC16, PCR conditions were as follows: a denaturation step of 2min at 95°C (3min for SC16), followed by 35 cycles at 95°C for 45s, 60°C for 45s (56°C for SC16) 72°C for 1min (2min for SC16) and a final step of 7min at 72°C. Primer pairs designed for these genes were SC9 F 5′-TAGGCGCGGTACTTGTTCAG-3′; SC9 R 5′-CTCCTCTGCGAAATCAATGCG-3′ and SC16 F 5′-GAAACTACAACTACAGTACTCC-3′; SC16 R 5′-AGCTCTGTTTCAGTGCATTTTC-3′. Amplification products were separated by agarose gel electrophoresis (1%) and visualized under UV by staining with ethidium bromide. Single band PCR products were sequenced using a 3730xl DNA analyzer (PE Applied Biosystems). Sequence assembly and edition was performed manually using CLC DNA Workbench (http://www.clcbio.com), and sequence alignments were performed using MUSCLE.7 DNA from additional isolates collected worldwide was used in this study for comparison with different strains in the Andean Colombian region. These samples were kindly donated by Michael Coffey (University of California, Riverside, USA).
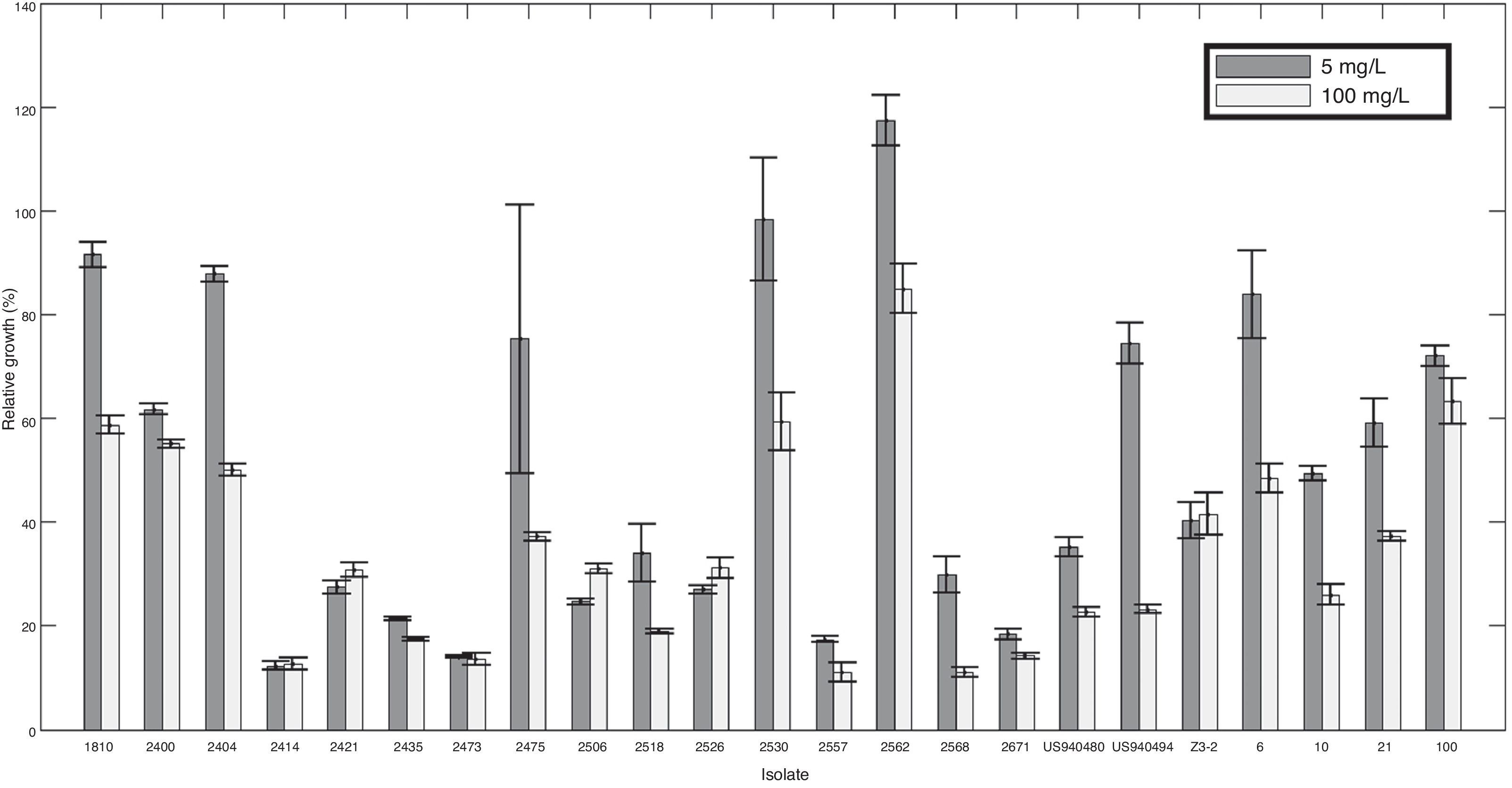
ResultsMefenoxam sensitivity tests evidenced resistanceForty-eight percent of the isolates showed some resistance level to mefenoxam (metalaxyl-M) (moderately resistant or resistant). Testing for resistance to this fungicide revealed that 52% of the isolates were sensitive, 22% intermediate and 26% resistant. Mefenoxam response was not associated with host origin, collection site or any of the characteristics such as mtDNA haplotypes, mating type or clonal lineage. At least one resistant isolate was found from each host sampled. No isolate showing intermediate resistance level was identified amongst strains from cape gooseberry plants, meanwhile the resistant isolate from this host had a higher average growth in the presence of mefenoxam (5mg/l) than in the medium without amendment (relative growth was 119%). This result was observed in two out of three repetitions of the test.
Two out of five resistant isolates were found in the region comprising the localities of Granada and San Francisco where no intermediate level was observed and sensitivity was most common compared to the overall population. All ranges of mefenoxam sensitivity were found in the Guasca region (Table 1). Isolate 2518 was the only strain completely inhibited at 100mg/l (Fig. 1).
Curzate and Ridomil Gold EC50 values for P. infestans isolatesMycelial sensitivity to Ridomil Gold and Curzate was tested and the effective concentration for which 50% of the mycelial growth was inhibited (EC50) to each fungicide was determined. EC50 values for Ridomil Gold ranged from 1.19 to 1.76mg/l with a mean EC50 value of 1.501mg/l. The Curzate EC50 values ranged from 1.13 to 2.04 with a mean EC50 value of 1.453mg/l. Initial concentrations tested for Ridomil Gold (2.5 and 6.3mg/l) inhibited 100% of the growth in all strains tested.
Virulence tests evidenced a high diversity of races in Central ColombiaAmongst the 20 strains tested, 19 physiological races were detected. In our sample, strains attacked an average of 4.3 of the 11 differentials tested (Table 1).
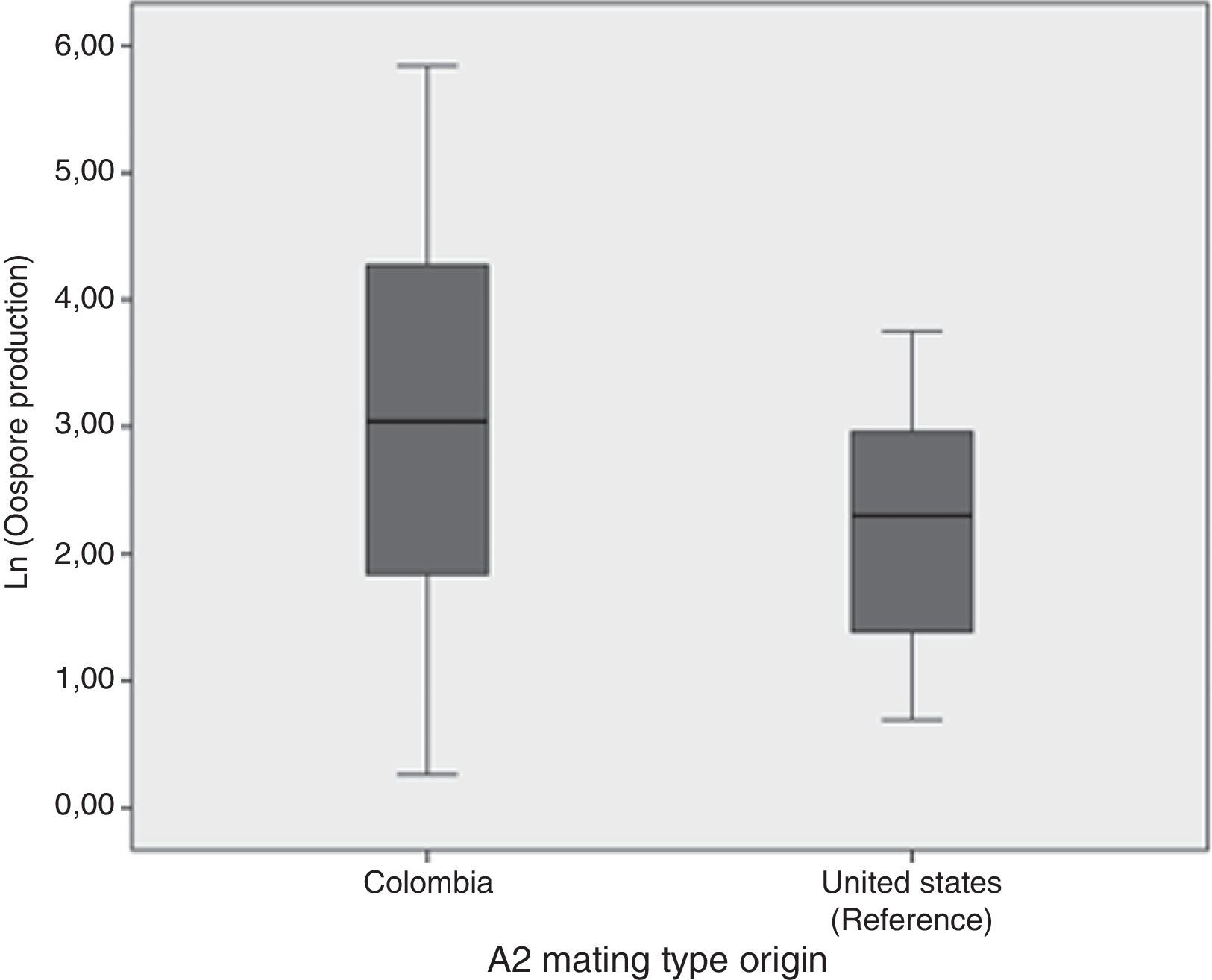
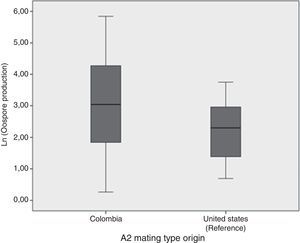
Oospore production assays showed the crossing potential of Colombian isolatesAll except two isolates tested (2404 and 2530) reproduced sexually in vitro when crossed with either the A2 reference isolate or the Colombian one. In general, oospore production was higher when Colombian A1 mating type isolate was crossed with the Colombian A2 mating type than when they were crossed with the A2 mating type reference isolate (Fig. 2). Nonetheless, according to the ANOVA results, the number of oospores produced did not depend on the A2 mating type strain used, Colombian or reference isolates (ANOVA: F=1.946; p=0.175). Additionally, a high level of variation in the number of oospores produced was observed amongst the Colombian A1 isolates. The isolate that showed the highest number of oospores was the 2435, isolated from S. tuberosum. The percentage of viability for the oospores produced ranged from 0.125% for the 2473 isolate to 26% for the 2562 isolate. No oospores were obtained for any isolate tested in either the in vivo assay or the soil assay even one month after the assay concluded.
Comparison of the natural logarithm (ln) of oospore production from 60 crosses between Colombian A1 mating types and the Colombian and reference (US94080) A2 isolate mating type. Oospore production was evaluated in vitro on a rye B agar plate. Crosses were kept at 18°C in the dark and they were checked for oospores production using a light microscope.
Using primers designed for the putative effector gene SC9 an amplification product length of 380bp was obtained. Only position 198 was polymorphic (A/G) among the strains tested in this study (GenBank accessions JN849408–JN849429) and the GenBank accession GQ869472.1. Using primers designed for the putative effector gene SC16 a PCR product of 330bp was obtained. Results of BLASTn (default parameters) showed that the Colombian SC16 sequence had 100% similarity and an e-value of 4e−33 with two putative RxLR effectors from P. infestans (GQ869431.1 and GQ869430.1). No polymorphisms were observed between any of our sequences and the GQ869430.1 sequence. Two nucleotides (positions 205 and 215) were polymorphic between our sequences and GQ869431.1 (data not shown).
No polymorphisms were found for the Avr3a gene (GenBank accessions JN849402–JN849407) (Fig. 3). For the β-tubulin gene (GenBank accessions JN849430–JN849450), no polymorphisms were obtained in the Colombian strains, except for strains 2414 and 2401 from Cogua that had the same nucleotide sequence as the United States reference strains. None of the obtained sequences corresponded to the previously reported haplotypes for the β-tubulin gene in the region4 (data not shown).
DiscussionThis is the first study performed on P. infestans in the central Andean Colombian region that includes both physiological and molecular approaches including the recently described RxLR effector gene sequences. A previous study that aimed at characterizing the pathogen population in the Northern Andean region4 highlighted the need to completely describe and understand local populations in Colombia as each region (Central and South) contained a unique group of strains. Our results demonstrated that isolates in this region show a high diversity of races, resistance to the most common fungicides used and monomorphism at the effector genes sequenced.
The most striking result of our study was the high diversity of races found in Central Colombia. The Colombian P. infestans population has been increasing race complexity through time.13,26 Race differentials are based on S. demissum, a wild relative of S. tuberosum, and in addition there are several wild Solanum species reported in Colombia, which may be P. infestans hosts.4 These findings may indicate that the pathogen is evolving to a wider host compatibility and may reflect the tremendous adaptability reported in several studies.
The low percentage of resistant strains in the Colombian Andean region to both, mefenoxam as technical grade (90%) and its commercial formulation in Ridomil Gold® (4%), is congruent with data previously reported by Garcia et al.,12 where the EC50 value for Ridomil Gold® was 0.62mg/l. Studies conducted in neighboring countries, such as Venezuela, showed that a high percentage of sensitive strains is common in the North Andean region.30 For curzate, we found an increase of effective concentration when compared with previous studies27,33 that suggests a shift in the level of fungicide resistance. All these results suggest that the fungicides tested still may be used in an integrated disease management program but correct fungicide application and continuous monitoring of the pathogen population is mandatory to keep these molecules useful for pathogen control and to decrease the risk of an outbreak of P. infestans resistant variants. In Colombia fungicides are used up to 15 applications per crop cycle.35 According to The Fungicide Resistance Action Committee (FRAC) spray of phenylamides such as mefenoxam must be limited to 1–2 consecutive applications per crop per cycle and not exceeding 14-day intervals.3 The detection of one individual with an enhanced growth on media supplemented with 50mg/l of mefenoxam in Colombia is not an exceptional event. Isolates growing on high concentrations of mefenoxam were reported in Canada and in the United States,38 and have also been reported in other species of Phytophthora such as P. capsici.25 This result reinforces the idea that a continuous monitoring of resistance to fungicides must be performed in P. infestans and the search for alternative control strategies should continue.
According to the characteristics described by Cooke and Lees in 20045 of the ideal molecular marker for assessing the diversity of P. infestans, effector genes could provide enough resolution to characterize a pathogen population due to the selection pressure exerted on these genes by the host. In addition, RxLR population sequence studies may provide evidence for host–pathogen co-evolution and RxLR gene sequence studies that include a high number of genes are very important to understand virulence mechanisms and pathogenicity. More than 500 RxLR genes have been identified from the P. infestans genome sequence19 but unfortunately, two new single copy RxLR genes selected in this study, did not help to discriminate the strains in the sample. Concerning Avr3a, although no polymorphisms were found, the strains tested in this study were similar to the haplotype 1 (H1) reported by Cárdenas et al.4 This haplotype differs from the allele sequences previously reported24 at nucleotide 139 causing a change in the amino acid sequence from methionine to isoleucine. The H1 haplotype has the same putative aminoacid sequences at positions 80 and 103 as the virulent Avr3a E80M103 allele (Fig. 3).1 Although the critical change reported to affect recognition by R3a is located at position 80 in the amino acid sequence,2 it is possible that the polymorphism identified in this study at position 139 also affects recognition. However, this has not been determined and hence no conclusions can be made on possible selection pressure exerted by the host either in commercial fields or in wild plants. Importantly, the polymorphism at position 132 is unique to Colombian strains and it could hence be used as a molecular marker to detect strain migrations.
The oospore assays showed that the Colombian strains have the possibility of sexual reproduction in vitro. However, the fact that in vivo or in soil the strains could not reproduce suggests that barriers to sexual reproduction could be extrinsic more than intrinsic. It is interesting that the same isolate that showed a higher number of viable oospores in its progeny was also resistant to mefenoxam. Nevertheless, this isolate was not the one that produced the highest number of total oospores. These results differ from those reported by Mukalazi et al.23 where the most resistant isolates were the ones producing the highest number of oospores. Here, the complete opposite behavior was observed and isolates that produced the highest number of oospores were mefenoxam-susceptible.
Even though the SC9 and SC16 effector genes did not show polymorphism to conduct a population genetic analysis, knowledge of the population structure using other effectors genes and response to different fungicides in the Colombian Andean region will be useful for the development of better disease management strategies including plant breeding and chemical control. Results shown in this study will contribute to the development of durable management strategies for this disease in the future. Over time, this and similar studies will provide information about changes in the population structure of this pathogen, which may help in the study of its evolutionary history, host interactions and contribute to a better understanding of its global population.
Conflict of interestThe authors declare that they have no conflict of interest.
Authors would like to thank La Facultad de Ciencias of the Universidad de los Andes for funding. We are also thankful to Dr. William Fry and Dr. Michael Coffey for kindly donating the reference strains used in this study.