Successful treatment of cryptococcosis requires rapid, accurate identification of Cryptococcus neoformans or the Cryptococcus gattii species complex in addition to their discrimination from other pathogenic yeast species. Gold nanoparticles (AuNPs) have enabled improvements in biomedical sciences owing to their versatile properties and capacity for detecting biological molecules at low concentrations; these methods are inexpensive, rapid, and stable.1,2,6–9 We describe the application of AuNPs for detecting C. gattii sensu lato.
Our protocol was approved by the Animal Research Ethics Committees (23108.014032/14-0) and Research Ethics Committees of Hospital Universitário Júlio Muller (888/CEP-HUJM/2010). Twenty-one clinical C. gattii sensu lato isolates and four C. neoformans sensu lato isolates from humans and animals were tested. DNA was extracted using the glass bead method3 and samples representing each of the eight major molecular types of the C. neoformans/gattii species complex were used (Table 1). DNA samples from non-Cryptococcus microorganisms served as controls (Table 1).
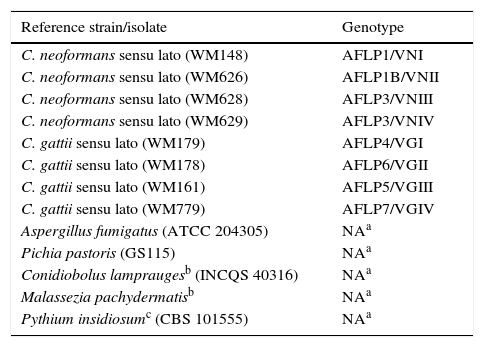
Reference strains representing each of the eight major molecular types of the Cryptococcus neoformans/gattii complex, identified using amplified fragment length polymorphism (AFLP) and restriction fragment length polymorphism (RFLP), and non-Cryptococcus strains.
| Reference strain/isolate | Genotype |
|---|---|
| C. neoformans sensu lato (WM148) | AFLP1/VNI |
| C. neoformans sensu lato (WM626) | AFLP1B/VNII |
| C. neoformans sensu lato (WM628) | AFLP3/VNIII |
| C. neoformans sensu lato (WM629) | AFLP3/VNIV |
| C. gattii sensu lato (WM179) | AFLP4/VGI |
| C. gattii sensu lato (WM178) | AFLP6/VGII |
| C. gattii sensu lato (WM161) | AFLP5/VGIII |
| C. gattii sensu lato (WM779) | AFLP7/VGIV |
| Aspergillus fumigatus (ATCC 204305) | NAa |
| Pichia pastoris (GS115) | NAa |
| Conidiobolus lampraugesb (INCQS 40316) | NAa |
| Malassezia pachydermatisb | NAa |
| Pythium insidiosumc (CBS 101555) | NAa |
AuNPs were synthesized using the citrate reduction method.4,5 Briefly, 250ml of 1mM gold chloride was boiled and added to 25ml of 38.8mM sodium citrate. For the hybridization test, we used probes based on the superoxide dismutase (SOD1) gene of C. gattii sensu lato (SOD1CGF 5′GATCCTCACGCCATTACG3′ and SOD1CGR 5′GAATGATGCGCTTAGTTGGA3′). First, 50ng of sample DNA was denatured at 95°C for 3min in a mixture containing 1.25M NaCl, 20mM Tris–HCl, and 20pmol oligonucleotides, followed by annealing at 50°C for 2min for hybridization and holding at 4°C. This was followed by addition of 50μl of the prepared AuNPs. Absorption (OD300–OD800) was measured using the BioTek® spectrophotometer (Winooski, VT, USA). The cutoff point was determined using the average absorbance values minus two standard deviations of five control samples without target DNA.
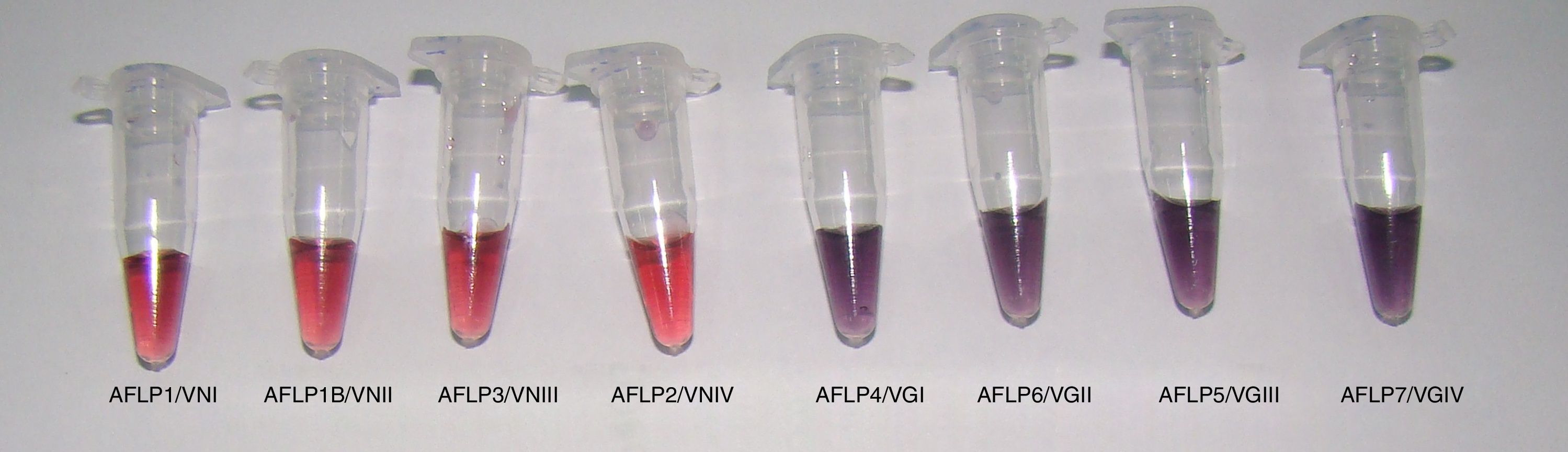
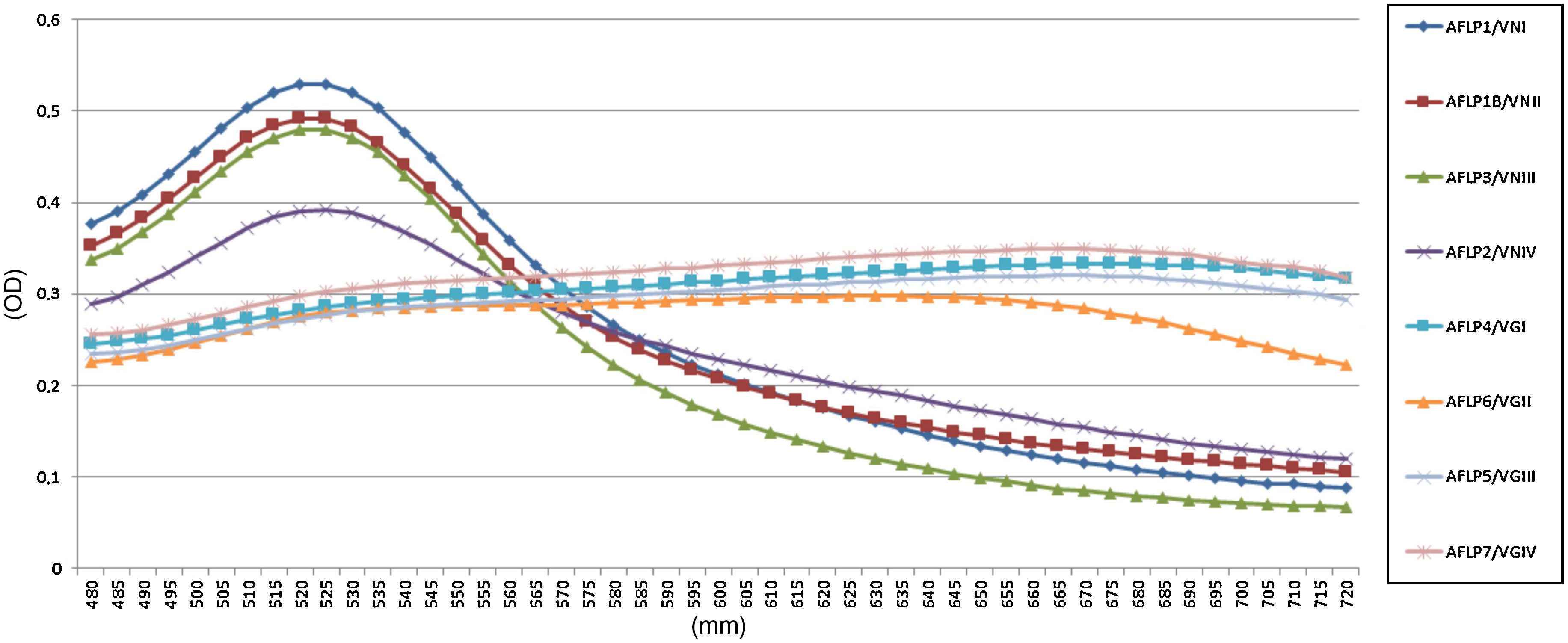
All reference samples were identified as C. gattii sensu lato, and a color change from red to purple indicated a positive reaction. Moreover, five non-Cryptococcus species and four C. neoformans sensu lato isolates were subjected to the AuNP assay to evaluate its specificity. All results were negative with no cross-reaction (100% specificity, Fig. 1).
In the AuNP test of the reference Cryptococcus strains, there were spectrophotometric signals at the 525 and 655nm absorbance peaks, indicating negative and positive results, respectively (Fig. 2). The average optical density at 525nm for the positive reference samples was 0.28 (confidence interval [CI]=0.27–0.30; SD=0.009), and the average for the negative samples was 0.47 (CI=0.39–0.52; SD=0.05). These results established that a positive sample yields an OD525≤0.37. Twenty-one (100%) clinical isolates of C. gattii sensu lato showed macroscopic positivity and OD525 values between 0.24 and 0.30, i.e., below the cutoff value for a negative result.
In summary, AuNPs serve as an alternative tool for developing new methods for clinical diagnosis; our AuNP-based test allowed rapid and specific identification of C. gattii sensu lato isolates. Its main advantages are its simplicity, ease of use, and low cost for set up without the need for sophisticated equipment.