The enzyme 3-hydroxy-3-methylglutaryl coenzyme-A reductase (HMGR) catalyzes the conversion of HMG-Co-A into mevalonate. This step is the limiting point for the synthesis of cholesterol in mammals and ergosterol in fungi. We describe in this article the genome organization of HMGR coding genes and those deduced from different fungi, recount the evidence showing statins as HMGR inhibitors for ergosterol synthesis and its effect in yeast viability, and propose fungal HMGR (HMGRf) as a model to study the use of pharmaceutical compounds to inhibit cholesterol and ergosterol synthesis. Bibliographical search and bioinformatic analyses were performed and discussed. HMGRfs belong to the class I with a high homology in the catalytic region. The sterol biosynthetic pathway in humans and fungi share many enzymes in the initial steps (such as the HMGR enzyme), but in the last steps enzymes are different rendering the two final products: cholesterol in mammals and ergosterol in fungi. With regards to inhibitors such as statins and other compounds, these affect also fungal viability. Since HMGR from Schizosaccharomyces pombe and Ustilago maydis are very similar to the human HMGR in the catalytic regions, we propose that fungal enzymes can be used to test inhibitors for a potential use in humans. We consider that HMGRf is a good therapeutic target to design and test new antifungal compounds.
This manuscript is part of the series of works presented at the “V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi” (Oaxaca, Mexico, 2012).
La enzima 3-hidroxi-3-metilglutaril coenzima A reductasa (HMGR) cataliza la conversión de HMG-Co-A a mevalonato, paso limitante en la síntesis de colesterol en mamíferos y de ergosterol en hongos. El presente artículo describe la organización de genes codificantes y proteínas de las diferentes HMGR de hongos (HMGRf), expone las evidencias disponibles en la inhibición de HMGR en la síntesis de ergosterol y su efecto en la viabilidad de los hongos, y propone las HMGRf como modelo de estudio para la aplicación de fármacos inhibidores de las síntesis de colesterol y ergosterol. Para ello se realizó una búsqueda bibliográfica y análisis bioinformáticos, con descripción de los datos. Las HMGRf son de clase i y presentan una alta homología en la región catalítica. La vía biosintética de esteroles en el ser humano y en los hongos comparte algunas enzimas iniciales (como la HMGR) pero, en los últimos pasos, las enzimas son diferentes, lo que genera productos finales distintos: colesterol y ergosterol, respectivamente. La inhibición de HMGRf por estatinas afecta a la síntesis de ergosterol y la viabilidad. Dado que el sitio catalítico de las HMGR de Schizosaccharomyces pombe y Ustilago maydis es muy similar al de la enzima humana, podrían servir como modelos para el estudio de fármacos inhibidores de la síntesis de colesterol. La HMGRf es una diana terapéutica adecuada para el diseño de nuevos antimicóticos.
Este artículo forma parte de una serie de estudios presentados en el «V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi» (Oaxaca, México, 2012).
The enzyme 3-hydroxy-3-methylglutaryl coenzyme-A reductase (HMGR) catalyzes the conversion of 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) to mevalonate, an oxido-reduction reaction that is a rate-limiting step in cholesterol synthesis in mammals, including humans, ergosterol in fungi, and other isoprenoids. The reaction catalyzed by HMGR is:
This enzyme is found in both eukaryotes (localized to the endoplasmic reticulum) and prokaryotes (soluble and cytoplasmic).11 Phylogenetic analysis has revealed two classes of HMGRs, class I enzymes grouping those from eukaryotes and some Archaea and class II enzymes grouping bacteria and certain other Archaea HMGRs.9 The reaction catalyzed by human HMGR is a target for hypocholesterolemic drugs such as statins, which are intended to lower cholesterol levels in serum.23 Furthermore, the HMGR enzyme of some opportunistic pathogenic fungi has been proposed as a target for inhibiting ergosterol synthesis as an alternative for solving the problem of the antifungal resistance.4,25 To this end, the effect of statins on HMGR inhibition in some fungi has been studied resulting in production of sterols, dolichol and coenzyme Q10 being affected.4,15,20,25
Despite the fact that HMGR activity in some yeasts such as Saccharomyces cerevisiae, Candida glabrata, Schizosaccharomyces pombe and others, has been described, there are no studies dealing with the genes encoding for this enzyme. Comparative analyses of putative proteins deduced from the nucleotide sequences encoding HMGRs have not been performed. Such analyses would provide relevant information to endorse the proposal of considering the HMGR enzyme as a therapeutic target. This review evaluates the conservation of the catalytic site HMGR from various fungi, its topology, activity inhibition and its possible use from to study HMGR inhibitors for human therapeutic use.
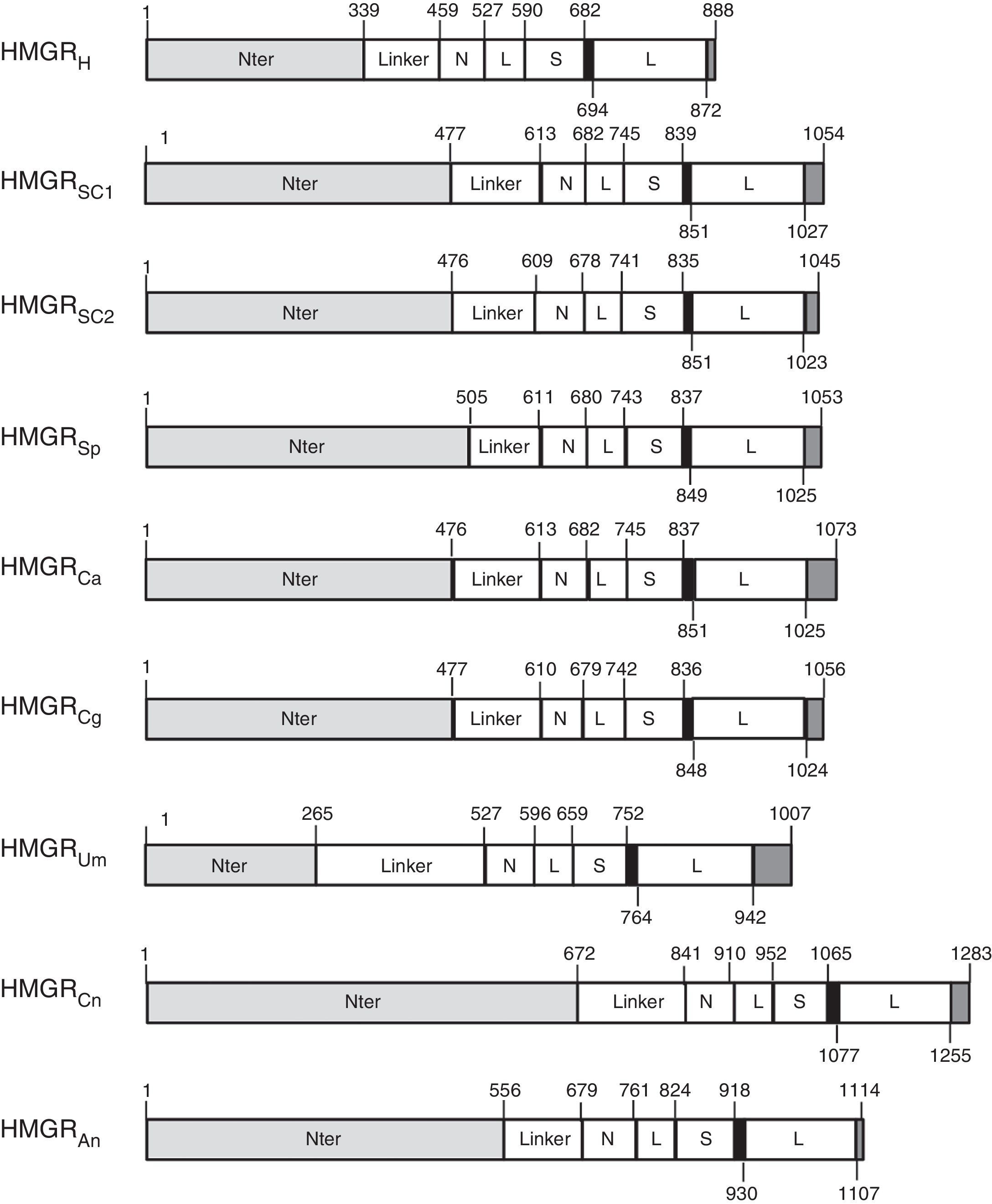
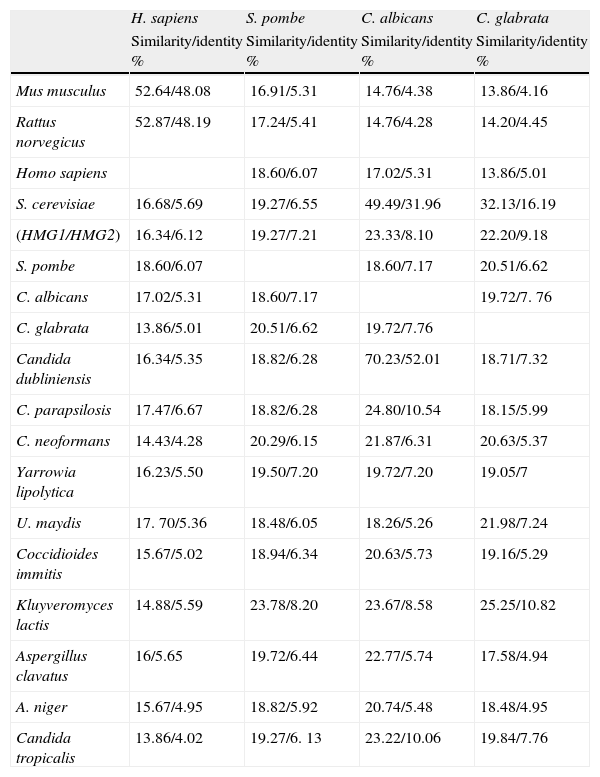
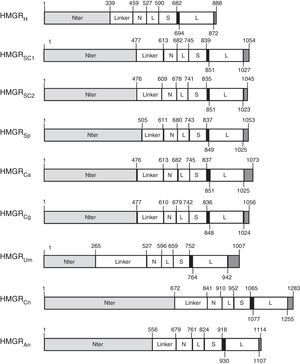
Fungal HMGR gene organizationAnalysis of genome sequences identified hmgr genes in organisms from all three domains of life. More than 150 HMGR sequences are recorded in public databases.9 The number of genes encoding the HMGR enzyme may depend on the organism or species. Animals, Archaea, and bacteria have only one hmgr gene, while plants exhibit multiple HMGR isoenzymes. S. cerevisiae has two HMGR isoenzymes (hmgr-1 and hmgr-2)2 while C. glabrata has a single gene (hmgr-1-Cg). In this review, we have identified HMGR sequences from S. cerevisiae (HMGRSC1 and HMGRSC2), S. pombe (HMGRSp), Candida albicans (HMGRCa), C. glabrata (HMGRCg), Ustilago maydis (HMGRUm), Cryptococcus neoformans (HMGRCn), and Aspegillus niger (HMGRAn), which were compared to human HMGR (HMGRH) (Fig. 1). Comparison of deduced aminoacid sequences confirms that analyzed fungal HMGRs belong to class I group. This latter means that all these enzymes are comprised of three characteristic domains: the membrane anchor domain, a linker and the catalytic domain. Some subdomains have been defined within the catalytic domain. The N domain connects the L domain to the linker domain; L domain contains an HMG-CoA binding region; and the S binds NADP(H)9 (Fig. 1). The HMGR enzymes from ascomycetes (Sc, Cg, An) and basidiomycetes (Um, Cn) show the three characteristic domains (Fig. 1). Analyzed proteins also have the amino acid sequence that binds the cofactor NADPH (indicated by a black box in Fig. 1, see residues 839–851aa from S. cerevisiae).9 Identity and similarity analyses of HMGR soluble fraction (i.e. the catalytic fraction) from multiple organisms, including different mammals and fungi, showed that all these catalytic domains are highly conserved (Table 1). When a comparison was carried out between HMGRs from different fungi and the human counterpart we observed that S. pombe, Candida parapsilosis and U. maydis presented the highest similarity with human HMGR (Table 1).
Schematic representation of HMGR proteins. Diagram shows features of HMGR proteins from: Human (HMGRH), S. cerevisiae (HMGRSC1 and HMGRSC2), S. pombe (HMGRSp), C. albicans (HMGRCa), C. glabrata (HMGRCg), U. maydis (HMGRUm), C. neoformans (HMGRCn), and A. niger (HMGRAn). Indicated domains are as follows: Nter, N-terminal domain containing a membrane anchor domain; Linker (connects membrane anchor domain to catalytic domain); the catalytic domain which has subdomains N, L, S, and L. N domain: connects L domain to linker; L domain: contains an HMG-CoA binding site; S domain: binds NADPH. Black box shows the cis-loop connects the HMG-CoA-binding region with the NADP-H-binding region.
Similarity and identity of multiple HMGRs class 1 from mammals and fungi.
| H. sapiens | S. pombe | C. albicans | C. glabrata | |
| Similarity/identity % | Similarity/identity % | Similarity/identity % | Similarity/identity % | |
| Mus musculus | 52.64/48.08 | 16.91/5.31 | 14.76/4.38 | 13.86/4.16 |
| Rattus norvegicus | 52.87/48.19 | 17.24/5.41 | 14.76/4.28 | 14.20/4.45 |
| Homo sapiens | 18.60/6.07 | 17.02/5.31 | 13.86/5.01 | |
| S. cerevisiae | 16.68/5.69 | 19.27/6.55 | 49.49/31.96 | 32.13/16.19 |
| (HMG1/HMG2) | 16.34/6.12 | 19.27/7.21 | 23.33/8.10 | 22.20/9.18 |
| S. pombe | 18.60/6.07 | 18.60/7.17 | 20.51/6.62 | |
| C. albicans | 17.02/5.31 | 18.60/7.17 | 19.72/7. 76 | |
| C. glabrata | 13.86/5.01 | 20.51/6.62 | 19.72/7.76 | |
| Candida dubliniensis | 16.34/5.35 | 18.82/6.28 | 70.23/52.01 | 18.71/7.32 |
| C. parapsilosis | 17.47/6.67 | 18.82/6.28 | 24.80/10.54 | 18.15/5.99 |
| C. neoformans | 14.43/4.28 | 20.29/6.15 | 21.87/6.31 | 20.63/5.37 |
| Yarrowia lipolytica | 16.23/5.50 | 19.50/7.20 | 19.72/7.20 | 19.05/7 |
| U. maydis | 17. 70/5.36 | 18.48/6.05 | 18.26/5.26 | 21.98/7.24 |
| Coccidioides immitis | 15.67/5.02 | 18.94/6.34 | 20.63/5.73 | 19.16/5.29 |
| Kluyveromyces lactis | 14.88/5.59 | 23.78/8.20 | 23.67/8.58 | 25.25/10.82 |
| Aspergillus clavatus | 16/5.65 | 19.72/6.44 | 22.77/5.74 | 17.58/4.94 |
| A. niger | 15.67/4.95 | 18.82/5.92 | 20.74/5.48 | 18.48/4.95 |
| Candida tropicalis | 13.86/4.02 | 19.27/6. 13 | 23.22/10.06 | 19.84/7.76 |
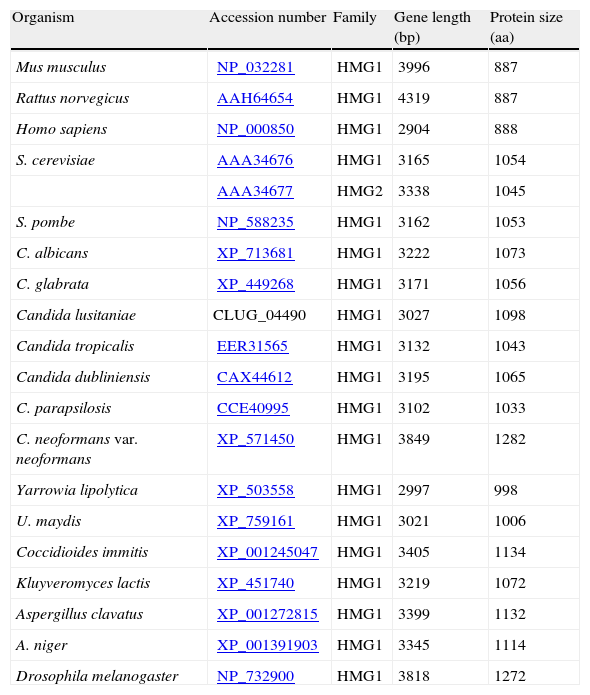
The analysis of the open reading frame (ORF) of different HMGRs showed that these can vary in size but sequences of all deduced proteins have the three characteristic HMGR domains from eukaryotic class I proteins: membrane anchor, linker and catalytic domains (Table 2). The analyses of the genes encoding different HMGRs, also demonstrated that have no introns (data not shown), unlike the genes encoding mammals HMGRs, which may contain up to 20 introns.9
Details of the sequences used for the structural protein analysis in Figs. 1 and 2 and identities/similarities in Table 1.
| Organism | Accession number | Family | Gene length (bp) | Protein size (aa) |
| Mus musculus | NP_032281 | HMG1 | 3996 | 887 |
| Rattus norvegicus | AAH64654 | HMG1 | 4319 | 887 |
| Homo sapiens | NP_000850 | HMG1 | 2904 | 888 |
| S. cerevisiae | AAA34676 | HMG1 | 3165 | 1054 |
| AAA34677 | HMG2 | 3338 | 1045 | |
| S. pombe | NP_588235 | HMG1 | 3162 | 1053 |
| C. albicans | XP_713681 | HMG1 | 3222 | 1073 |
| C. glabrata | XP_449268 | HMG1 | 3171 | 1056 |
| Candida lusitaniae | CLUG_04490 | HMG1 | 3027 | 1098 |
| Candida tropicalis | EER31565 | HMG1 | 3132 | 1043 |
| Candida dubliniensis | CAX44612 | HMG1 | 3195 | 1065 |
| C. parapsilosis | CCE40995 | HMG1 | 3102 | 1033 |
| C. neoformans var. neoformans | XP_571450 | HMG1 | 3849 | 1282 |
| Yarrowia lipolytica | XP_503558 | HMG1 | 2997 | 998 |
| U. maydis | XP_759161 | HMG1 | 3021 | 1006 |
| Coccidioides immitis | XP_001245047 | HMG1 | 3405 | 1134 |
| Kluyveromyces lactis | XP_451740 | HMG1 | 3219 | 1072 |
| Aspergillus clavatus | XP_001272815 | HMG1 | 3399 | 1132 |
| A. niger | XP_001391903 | HMG1 | 3345 | 1114 |
| Drosophila melanogaster | NP_732900 | HMG1 | 3818 | 1272 |
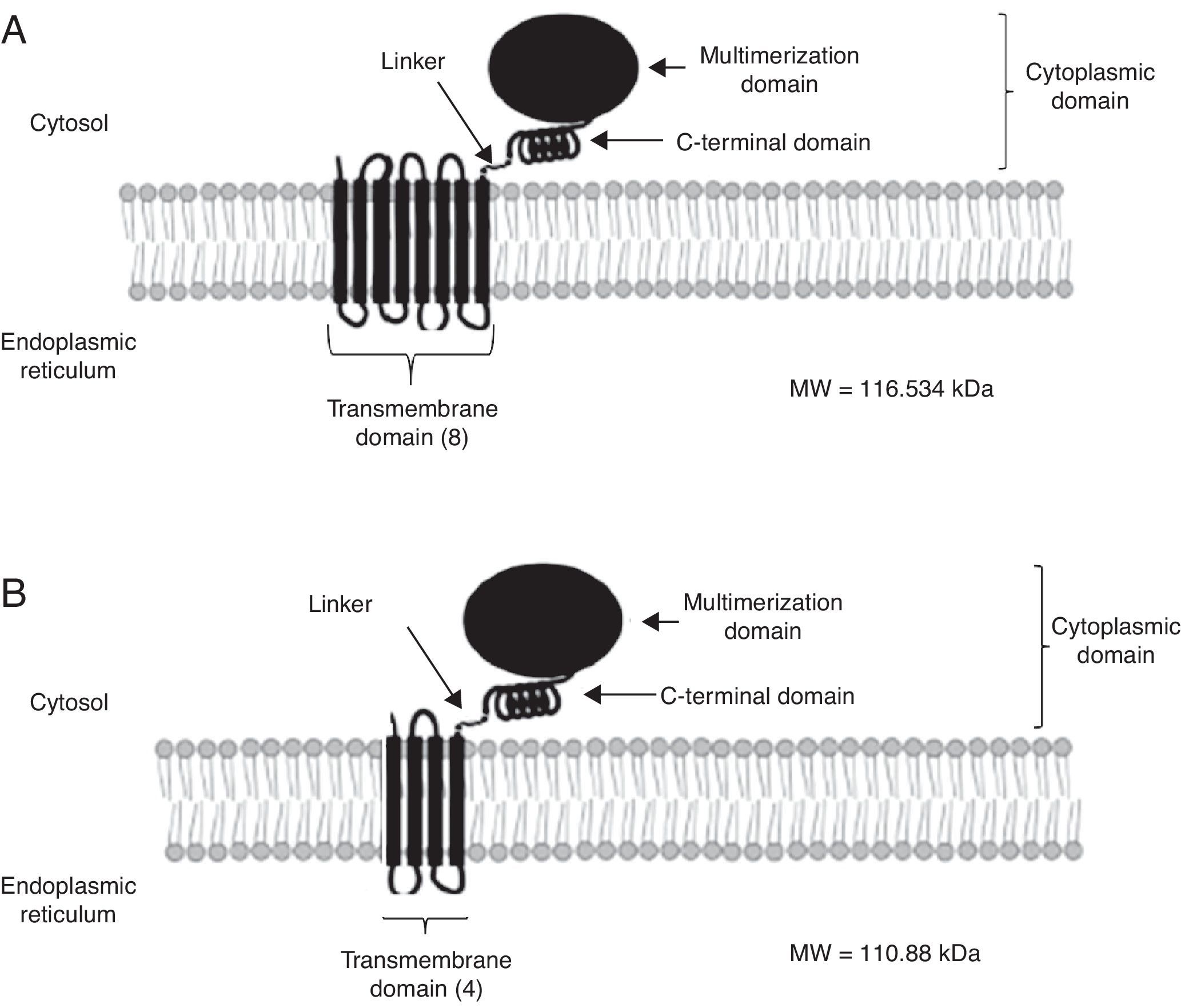
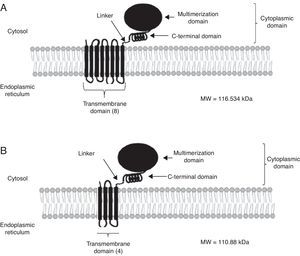
High-resolution crystal structures have been solved for class I human HMGR (HMGRH).12 The crystal structure shows the oligomeric state of this enzyme and suggests relevance for activity and a mechanism for cholesterol sensing. The active site architecture of human HMGR is different from that of bacterial HMGR and this may explain why HMGR inhibitors in bacteria have not been reported. Owing to the fact that the active site domain of S. pombe HMGR is quite similar to that of humans, a new approach has been developed by using the fungal HMGR (HMGRf) as a straight forward in vitro assay for measuring inhibitory activity of the test compounds designed as hypocholesterolemic in vivo in a murine model.1 Although the fungal enzymes examined in this review showed the characteristic domains of the class I enzymes (transmembranal, linker and catalytic), certain differences can be observed in the protein topology. For instance, C. glabrata HMGR (HMGRCg) has eigth possible transmembrane sequences while U. maydis HMGRUm has only four (Fig. 2). This suggests a different regulation between both enzymes. As a matter of fact, it has been suggested that the transmembrane region participates in HMGR degradation (post-translational regulation).9 HMGRH was purified as a homotetramer and the predicted topology of HMGRCg and HMGRUm shows multimerization domains too, suggesting that these may also form multimers (Fig. 2).
Model for the secondary structure of Candida glabrata HMGR (HMGRCg) and Ustilago maydis HMGR (HMGRUm). Secondary structure was determined from HMGRCg (A) and HMGRUm (B). Both enzymes show some characteristics of other eukaryotic HMGRs, this is: an amino terminus facing the lumen of the endoplasmic reticulum and the carboxyl terminus facing the cytoplasm, eighth or four transmembrane segments (C. glabrata and U. maydis, respectively), and a multimerization domain which forms the binding domain for the substrate and the cofactor NADPH.
As we mentioned before, yeasts produce ergosterol using a metabolic pathway that in many enzymatic steps is similar to that found in mammals, with HMGR being the limiting step. With such a relevant role in cell physiology, this enzyme is tightly regulated at different levels including transcriptional, post-transcriptional, traductional and post-traductional regulation.14 Interestingly, the enzymatic activity of HMGR does not produce toxic precursors and therefore it is an attractive target for controlling cholesterol levels in humans.23 After the discovery of compactin in 1970, a specific HMGR inhibitor, in growing cultures of Penicillium citrinum,8 many other models have been studied, including rabbits, non-human primates, rats and dogs, to detect changes in cholesterol levels. In 1978 the Aspergillus terreus HMGR was used as a model to study the inhibitory effect of mevinolin, later called lovastatin.23 These and other studies served for the classification of inhibitors in two groups: analogs of the 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) and those that are not, such as alpha-asarone-based analogs.22 Despite the fact that multicellular organisms models are attractive because of their similarity with humans they have proven to be difficult to work with. Therefore, the development of new unicellular models has become an attractive option. For instance, the yeast S. pombe has been used to study the regulatory aspects of sterol biosynthesis.1
The ergosterol biosynthetic pathway is fully known in the yeast S. cerevisiae.16 It is composed of 20 enzymatic reactions and it is divided into the mevalonate pathway, which includes 9 reactions and ends with the synthesis of farnesyl pyrophosphate, and the pathway that converts this intermediate into ergosterol with the involvement of 11 enzymatic reactions.7 This pathway is responsible for the biosynthesis of important intermediate products such as the geranylgeraniol group, which is involved in protein post-traductional modifications (i.e. Ras, Rac), dolichol, heme group and quinolone synthesis.7,10 Experimental evidence has shown that synthesis of ergosterol in S. pombe follows a similar pathway to that of S. cerevisiae.3,13
Inhibition of viability and ergosterol synthesis in yeasts as a consequence of the inhibition of the HMGRUp to date there have been published some studies about the effect of statins, azoles or their combination against fungi in liquid or solid medium to determine antifungal activity. Accordingly, it has been observed that lovastatin is capable of affecting sterol levels in S. cerevisiae.18 Statins may also inhibit the growth of Candida species and Aspergillus fumigatus.21 Other studies demonstrated synergism of azoles with statins, such as fluvastatin with fluconazole or itraconazole which together, can reduce ergosterol levels and viability in Candida spp. and C. neoformans.5 Similarly it has been demonstrated that the synergism of statin and azoles inhibit ergosterol synthesis in S. cerevisiae and Candida utilis in a bioassay on solid medium.4 On the other hand, a series of alpha-asarone and fibrate-based analogs were designed by docking approaches with published crystal structures of human HMGR and the partial purification of the enzyme from S. pombe allowed the test of analog compounds, resulting in positive and significant inhibitory activity.1,24 The use of simvastatin in C. glabrata has shown that in addition of reducing the levels of ergosterol and inhibiting growth, this statin can lead to the loss of mitochondrial DNA (mtDNA).25 This may restrict the use of statins as antifungal agents, because, the loss of mtDNA and the generation of “petite” mutants are related to azole resistance in C. glabrata. Therefore, new synthetic compounds are being designed, analogs of statins, capable of inhibiting the synthesis of ergosterol but with no loss of mtDNA (data not shown).
HMGR enzyme as model to identify novel therapies to modulate cholesterol synthesis and to study sterol biosynthesisAlthough yeasts synthesize ergosterol instead of cholesterol, fungi share many of the enzymes involved in the sterol synthesis pathway in mammals, including HMGR. Therefore, the fungal HMGR have proven to be suitable models for studying the inhibition by compounds structurally related to statins or fibrates. In this sense, the enzyme from S. pombe has been proposed as a model to study some aspects of regulation of sterol biosynthesis that have been difficult to address in other organisms.1,17,20 Heterologous expression of genes encoding the HMGR from different fungi also has been successful to study pharmaceutical compounds inhibiting cholesterol synthesis. Accordingly, the Rhizomucor miehei HMGR gene, has been expressed in Mucor circinelloides.19 HMGR C-terminal from U. maydis was expressed in Escherichia coli and shown to be blocked by a potent inhibitor of mammalian and fungal HMG-CoA reductases.6
ConclusionsGiven the similarities in the initial steps of cholesterol and ergosterol synthesis pathways, the fact that HMGR is a critical control point in both of them and that the enzyme from S. pombe and U. maydis are very similar to the human counterpart in the catalytic regions, we propose that fungal enzymes can be used to test inhibitors with a potential use in humans.
Conflict of interestThe authors declare that they have no conflict of interests.
We would like to thank Cesar I. Ortíz-García for assisting us preparing material for this review. DAP, ESS, and BRA, are CONACyT fellowships, DAP is PIFI fellowship, and grants from CONACyT 133695 and SIP-IPN-20131171 were received. JT, CHR and LVT are EDI and COFAA fellowships. JAI was hired by program “Contratación de investigadores para el apoyo a la investigación y posgrado-IPN”.