Candida glabrata, a haploid and opportunistic fungal pathogen that has not known sexual cycle, has conserved the majority of the genes required for mating and cell type identity. The C. glabrata genome contains three mating-type-like loci called MTL1, MTL2 and MTL3. The three loci encode putative transcription factors, a1, α1 and α2 that regulate cell type identity and sexual reproduction in other fungi like the closely related Saccharomyces cerevisiae. MTL1 can contain either a or α information. MTL2, which contains a information and MTL3 with α information, are relatively close to two telomeres. MTL1 and MTL2 are transcriptionally active, while MTL3 is subject to an incomplete silencing nucleated at the telomere that depends on the silencing proteins Sir2, Sir3, Sir4, yKu70/80, Rif1, Rap1 and Sum1. C. glabrata does not seem to maintain cell type identity, as cell type-specific genes are expressed regardless of the type (or even absence) of mating information. These data highlight important differences in the control of mating and cell type identity between the non-pathogenic yeast S. cerevisiae and C. glabrata, which might explain the absence of a sexual cycle in C. glabrata. The fact that C. glabrata has conserved the vast majority of the genes involved in mating might suggest that some of these genes perhaps have been rewired to control other processes important for the survival inside the host as a commensal or as a human pathogen.
This manuscript is part of the series of works presented at the “V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi” (Oaxaca, Mexico, 2012).
Candida glabrata, una levadura patógena haploide y oportunista, que carece de ciclo sexual conocido (asexual), conserva la mayoría de genes ortólogos requeridos en los procesos de apareamiento, esporulación y la identidad del tipo celular. El genoma de C. glabrata contiene 3 loci de apareamiento llamados MTL1, MTL2 y MTL3 que codifican los presuntos factores de transcripción a1, α1 y α2 que controlan la reproducción sexual e identidad celular en otros hongos, como Saccharomyces cerevisiae con el cual tiene una estrecha relación filogenética. MTL1 puede contener información a o α; MTL2 contiene información a, y MTL3 que contiene información α1 y α2 son loci próximos a 2 telómeros. MTL1 y MTL2 son activos transcripcionalmente mientras que MTL3 está sujeto a un silenciamiento que no es completo, que proviene del telómero y depende de las proteínas Sir2, Sir3, Sir4, yKu70/80, Rif1, Rap1 y Sum1. C. glabrata parece no mantener identidad de tipo celular ya que varios genes específicos de un tipo celular se expresan en todas las células con independencia del tipo de información de apareamiento en los loci MTL, o incluso, en su ausencia. Estos datos ilustran varias diferencias importantes entre la levadura no patógena S. cerevisiae y C. glabrata que podrían explicar la característica asexual en esta última. El hecho de que en C. glabrata se hayan conservado los genes necesarios para el apareamiento podría indicar que es posible que algunos de estos genes se hayan «reorganizado» para controlar otros procesos importantes en la supervivencia de C. glabrata en su huésped, como comensal o como patógeno.
Este artículo forma parte de una serie de estudios presentados en el «V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi» (Oaxaca, México, 2012).
Sexual reproduction is thought to be advantageous in spite of the high cost in energy associated with it. Sex is thought to provide a means to eliminate deleterious mutations and promote genetic recombination, which in turn might produce progeny with a combination of beneficial traits that may be better adapted to changing environmental conditions.26 Indeed, recent experiments in the model yeast Saccharomyces cerevisiae support this idea. The data indicate that under stressful conditions, sexual cells have an advantage over the obligate asexual congenic strain that only differed in their respective ability to mate. Instead, when these strains were grown under mild non-stressful conditions, there was no advantage for the obligate sexual strain.13 Sexual reproduction is widely spread among eukaryotic organisms, even in many microbes that can reproduce asexually. However, there are some eukaryotes, including some fungi that appear to lack a sexual cycle. Only a few of the known species of fungi are associated with human disease, and many of the human pathogens have long since been thought to be asexual since no sexual reproduction has been observed. It is thought that sex in these pathogens might lead to loss of particular combinations of genes required to survive within the host.
Examples of asexual fungal pathogens are Sporothrix schenckii,29Coccidioides immitis, Coccidioides posadasii,11Candida parapsilosis,40Candida glabrata and others. However, in recent years there has been a surge in genomic studies that show that several of the human fungal pathogens contain the genes required for sexual reproduction, suggesting that they may have a cryptic sexual cycle, one that is controlled by very specific conditions, reviewed in some bibliographic references in this article.21,24,32,33 This is the case of C. immitis, C. posadasii,11Candida tropicalis,40Aspergillus fumigatus,37Candida albicans17 and C. glabrata.9,44 For some of these fungal pathogens, a cryptic sexual or parasexual cycle was later discovered, like the case of C. albicans,18C. tropicalis,38A. fumigatus34 and Paracoccidiodes brasiliensis.42 These pathogens reproduce sexually under very particular conditions. In the case of C. albicans and C. tropicalis, in order to mate cells first have to undergo a morphological switch, which is the mating-competent stage.30 For A. fumigatus, the sexual cycle can be observed in the laboratory after prolonged incubation (over 6 months) in the dark.34 In Cryptococcus neoformans even though a sexual cycle has been documented in the laboratory for a long time,22 mating is limited by the almost unisexual geographic distribution of only the α mating type.25 In this way, these pathogens have retained the ability to generate genetic variation through controlled sexual or parasexual cycles in response to specific or changing conditions.2,33
For sexual reproduction to occur, two cells of opposite mating type must recognize each other through mating-specific pheromone and receptor signals. This is followed by cell–cell fusion, nuclear fusion and in many fungi by meiosis, although in some fungi like C. albicans, no meiosis has been observed, instead mating products undergo gradual chromosome loss until haploid chromosome content is achieved.1
In this review we will focus on the common opportunistic pathogen C. glabrata, an asexual haploid yeast, that shares a closer phylogenetic relationship with S. cerevisiae than to other Candida species.5,14C. glabrata is a commensal in healthy individuals but can become a successful pathogen associated with high mortality rates in immunocompromised patients.36
Control of sexual reproduction and cell type identitySexual reproduction and cell-type identity in most fungi are controlled by the genes encoded in the mating type locus called MAT (or MTL in some fungi). This locus encodes transcription factors that regulate the expression of genes that determine cell type identity and the signaling cascades that enable the cell to respond to the pheromone secreted by cells of the opposite mating type. These transcription factors are usually proteins containing a homeodomain or other types of regulatory domains like a α-domain or HMG (high mobility group) domain. Early studies in the non-pathogenic yeast S. cerevisiae have led to a detailed molecular mechanism for cell type identity control and mating. This organism, which can reproduce both sexually and asexually, contains three mating type loci (MAT, HML and HMR) of which only MAT is transcriptionally active and the other two loci are maintained repressed by a mechanism known as silencing.15 Information present at the MAT locus can be either a-type or α-type while HML and HMR contain α and a information respectively, in over 97% of the strains studied. Control of cell type and mating involves a regulatory circuit determined by the a1 protein and the α1 and α2 proteins encoded in the MATa and MATα loci, respectively. The a1 and α2 genes encode homeodomain-containing transcription factors while the α1 gene encodes a protein containing a α domain. In MATa haploids only a-specific genes (asg) are expressed whereas in MATα haploids only α-specific genes (αsg) are expressed. In both types of haploids a set of genes specific for haploid cells (hsg) is also expressed. After mating, the resultant diploid (a/α) forms a heterodimer with the proteins a1 and α2 that represses α1, the hsg and some other genes involved with certain types of stress.12,15,19 This regulatory circuit with some modifications also controls cell type identity in C. albicans, a diploid opportunistic human pathogen. Notably, a heterodimer composed of a1/α2 proteins is also formed in this organism and negatively regulates the phenotypic switch required for mating, therefore, indirectly controlling mating.30
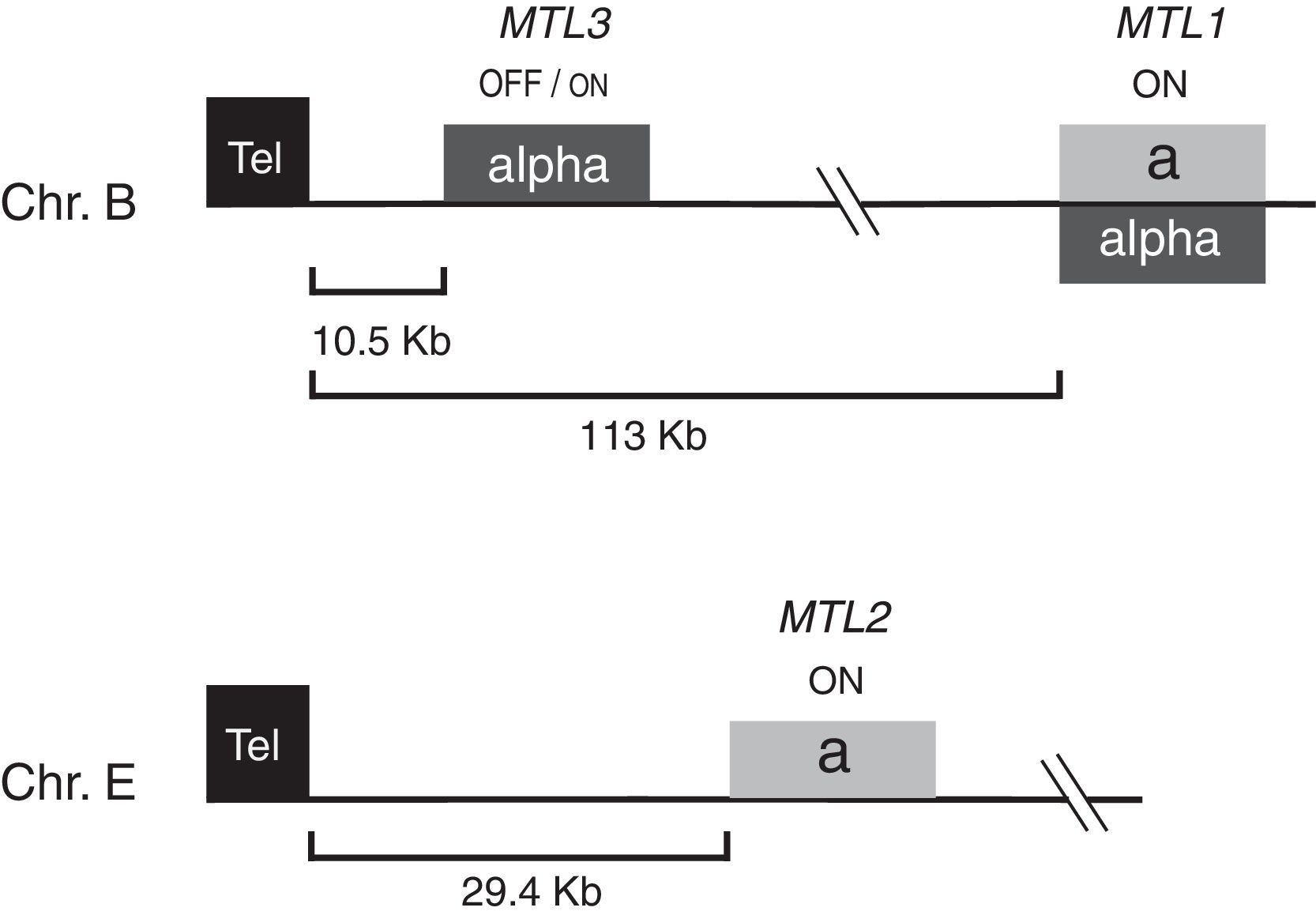
Structure of the C. glabrata mating type-like loci MTL1, MTL2, and MTL3Early studies reported that C. glabrata contains three mating type-like loci (MTL), in a similar configuration to that of the MAT, HML and HMR loci in S. cerevisiae.41MTL1 corresponds to MAT while MTL2 and MTL3 to HMR and HML, respectively. In C. glabrata, MTL1 and MTL3 are in chromosome B, and MTL2 is in chromosome E, whereas in S. cerevisiae the three loci are in chromosome III. It was initially proposed that MTL1 is the expression locus and that MTL2 and MTL3 are transcriptionally silent, analogous to the situation in S. cerevisiae.41 In the vast majority of C. glabrata isolates (approximately 97%), the information at MTL2 is a, and α in MTL3. In contrast, the information in MTL1 can be of either type, for example in the sequenced strain CBS138 (http://www.genolevures.org/cagl.html#), MTL1 contains α whereas the BG14 strain7 contains a information in this locus.39 It has been reported that there is a bias toward a-type of information at MTL1 in a collection of 190 C. glabrata clinical isolates from Africa, Europe, North America and South America where it was found that approximately 80% of the isolates contain a information and only 20% are α at this locus.4 However, in our collection of 79 clinical isolates from three hospitals in Mexico, the α containing isolates are more frequent, since 64% of isolates contain α information at MTL1 and 36% contain a information23 (and Robledo-Márquez and Castaño, unpublished data). Therefore, it seems that the distribution of mating types varies depending on the geographical sites where the C. glabrata isolates are collected. It is not known whether there is a correlation between the information at MTL1 and the pathogenicity in C. glabrata as there seems to be in C. neoformans where the α strains are more virulent than the very uncommon a strains.10,25
MTL1 and MTL2 are both transcriptionally activeMTL1 is at an internal position on chromosome B (113kb from the left telomere); MTL2 and MTL3 are both close to two telomeres, MTL2 is 29.4kb from the left telomere on chromosome E and MTL3 is only 10.5kb from the left telomere on chromosome B (Fig. 1). To determine whether MTL1 is expressed and MTL2 and MTL3 loci are silenced as suggested initially,41 we introduced at least 5 independent URA3 gene reporter insertions in each of the three loci and measured its expression using a plate growth assay on SC plates with 5-FOA, which is toxic to cells expressing URA3. Surprisingly, we found that MTL2 is in a conformation active for transcription as all the reporter insertions in this locus were expressed giving rise to Ura+, 5-FOAS strains. As expected, MTL1 is also transcriptionally active and cells of all reporter strains in this locus are also Ura+, 5-FOAS.39 Muller et al.31 also reported expression from both MTL1 and MTL2 in C. glabrata as measured by quantitative RT-PCR. The a1 gene contains two introns, and interestingly, the correct processing of the a1 mRNA only occurs when the transcript originates from the MTL1 locus, but not when transcription starts at MTL2.31,39 It is not known yet, what are the signals that direct processing of this mRNA since the sequence of the gene and the immediate flanking sequences are identical in the two loci (at least 370bp on either the 5′ or 3′ end), although it is possible that relative transcription rates might regulate this process through a chromatin based mechanism.28 The locus-specific processing of the a1 transcript was thought to be a mechanism to maintain cell type identity. However, by using strains with different combinations of knock-out mutations in the MTL loci, including a strain with no mating information [(mtl1,2,3)Δ], we found that several genes that are regulated in a cell type-specific manner in S. cerevisiae, are expressed in all strains in C. glabrata regardless of the information contained in the MTL loci and even in the absence of these loci. This suggests that C. glabrata might not maintain a cell type identity.39
Structure of the MTL loci of Candida glabrata. C. glabrata contains three mating type like loci (MTL). MTL1 is located in chromosome B at an internal position and can contain mating information type a or type α as indicated. MTL2 localized at 29.4kb from the left telomere of chromosome E generally contains a information and MTL3 which is localized at 10.5kb from the left telomere of chromosome B usually contains α information. MTL1 and MTL2 are transcriptionally active (indicated by ON) and MTL3 is subject to incomplete silencing spreading from the telomere (indicated by OFF/on).
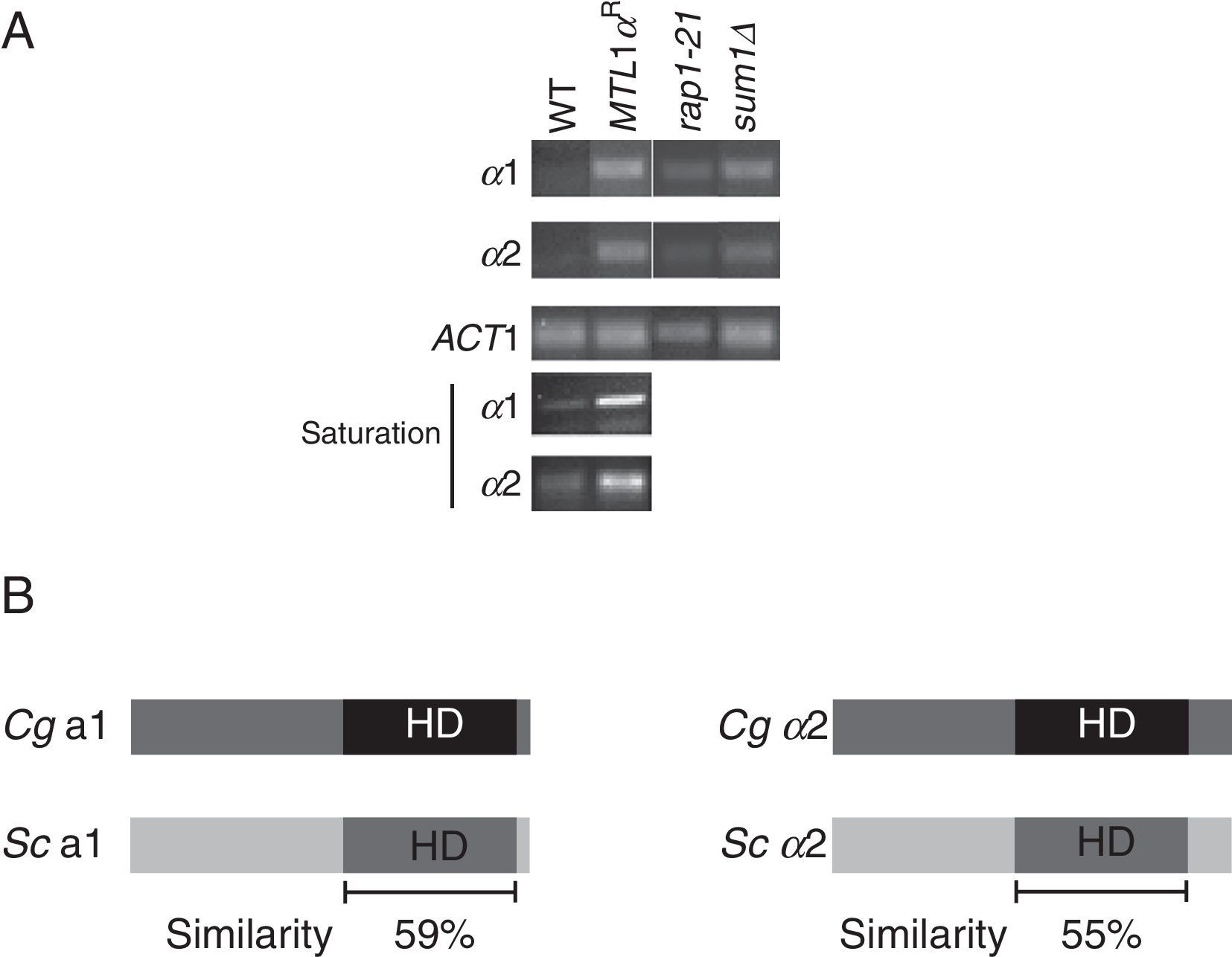
The MTL3 locus is the only MTL that is not transcriptionally active since in strains carrying reporter insertions throughout the locus, a large proportion of cells were able to grow on plates containing 5-FOA. This silencing is not complete because in every reporter insertion tested, we also found a population of cells that expressed the URA3 gene and were able to grow on plates without uracil.39 Even though there is no detectable expression of the native α1 and α2 genes present at MTL3 by RT-PCR,31,39 however, when the PCR is made under saturation conditions, a transcript is detected for both genes (Fig. 2A). These results are in contrast to the situation in S. cerevisiae where the two loci HMR and HML are very efficiently silenced through the activity of two cis-acting silencers flanking each locus where the silencing proteins Abf1, ORC and Rap1 bind.3,16,20
Subtelomeric silencing of MTL3 depends on Rap1 and Sum1 proteins. (A) RT-PCR of the α1 and α2 genes from MTL3 from the wild-type (wt), the rap1-21 or sum1Δ strains as indicated, or from MTL1 in the mtl(1, 2, 3)Δ strain where α1 and or α2 were reconstituted at MTL1 (MTL1 αR). The bottom part of the figure shows the RT-PCR performed at saturation conditions. RT-PCR for ACT1 gene was used as loading control. (B) Schematic representation and comparison of the a1 and α2 genes from S. cerevisiae and C. glabrata. HD represents the homeodomains of each protein and the similarity in this domain for each pair of proteins is indicated as percentage.
Silencing at MTL3 of C. glabrata was shown to be dependent on the Sir2, Sir3 and Sir4 proteins as well as Rif1 and yKu70 and yKu80 proteins; these data also differ markedly from the silencing of the HM loci of S. cerevisiae where yKu70 and yKu80 are not required for silencing, and only when SIR1 is deleted, a modest effect of the yKu proteins is observed.35,43 The silenced chromatin structure is probably nucleated at the telomere (and not from flanking cis-acting silencers), and spreads over 11kb to MTL3, because moving the entire MTL3 locus to an internal position in chromosome L results in expression of both, the URA3 reporter insertions and the α1 and α2 genes at that location.39 We have also recently shown that silencing at MTL3 also depends on Rap1, by using a strain containing allele rap1-21 (a deletion of the 28 carboxy-terminal amino acids of the protein), which is defective for subtelomeric silencing at various telomeres in C. glabrata.6,8 In the rap1-21 strain, but not in the wild-type strain, transcription of the α genes from MTL3 can be detected (Fig. 2A), supporting the idea that silencing at MTL3 comes from the telomere.
The transcriptional repressor Sum1 was discovered as an additional negative regulator of α-specific gene expression in MATa cells in Saccharomyces bayanus and S. cerevisiae. Repression by Sum1 in both organisms is important to prevent mating as α cells in MATa cells.45 The C. glabrata and the S. cerevisiae Sum1 proteins are only 20% similar over their entire lengths. To investigate whether Sum1 in C. glabrata regulates α1 and α2 expression, we measured α1 and α2 expression in a sum1Δ strain by RT-PCR. Fig. 2A shows that a deletion of Sum1 in C. glabrata results in de-repression of α1 and α2 genes from MTL3, suggesting that Sum1 represses the α genes at MTL3.
The fact that in C. glabrata silencing at the MTL3 locus is not complete might mean that in any given culture of strains of C. glabrata containing MTL1a and MTL3α, a small proportion of cells in the population could express both types of information and therefore a heterodimer between a1 and α2 could be formed in an analogous way to S. cerevisiae. This could result in the negative regulation of some genes if this putative heterodimer acts in a similar way to S. cerevisiae. In S. cerevisiae, the homeodomain of a1 interacts with 16 amino acids in the carboxy-terminal of the α2 protein.27 The a1 proteins from C. glabrata and S. cerevisiae are 46% homologous across the entire protein, and in the homeodomain, where it interacts with α2, they are 59% homologous of which 32% are identities (Fig. 2B). In the case of α2, the C. glabrata and the S. cerevisiae proteins are 56% homologous and at the 16 amino acids in the C-terminal tail where the interaction with a1 takes place, they are 43% homologous (of which 25% are identical). Therefore, it is possible that when both types of mating information are expressed in C. glabrata a heterodimer could be formed; we are investigating this possibility.
From the data discussed, it is clear that there are important differences in the expression of the cell type-specific genes and the genes encoded in the MTL loci between C. glabrata and S. cerevisiae that might explain the absence of a sexual cycle in C. glabrata. However, the fact that the genes that are required for mating and cell type identity are conserved in C. glabrata could mean that some of these genes might have been rewired to control the expression of a different set of genes or perhaps participate in some process that is important to the survival within the host, either as a commensal or as a pathogen. Alternatively, C. glabrata might have a cryptic sexual cycle regulated by an as yet unknown mechanism. Further studies on the expression of cell type specific genes and pheromone response pathway genes will help clarify whether some of these genes control processes that are important to the survival in the host.
Conflict of interestThe authors have nothing to declare.
This work was supported by grant No. CB2010-01-151517 to I.C.N. P.Y.C. was supported by fellowship No. 270403 and K.A.R.M. by fellowship No. 375866.