For many years fluconazole has been commonly used to treat Candida infections. However, the indiscriminate use of this antimycotic therapy has favored the emergence of resistant isolates. Mutations in the ERG11 gene have been described as one of the primary mechanisms of resistance in Candida species.
AimsIn this study we investigated missense mutations in ERG11 genes of Candida albicans, Candida glabrata and Candida tropicalis isolates previously evaluated by susceptibility testing to fluconazole.
MethodsScreening for these mutations was performed on 19 Candida clinical isolates (eight C. albicans, five C. glabrata and six C. tropicalis) resistant and susceptible to fluconazole. The ERG11 gene was amplified by PCR with specific primers for each Candida species and analyzed by automated sequencing.
ResultsWe identified 14 different missense mutations, five of which had not been described previously. Among them, a new mutation L321F was identified in a fluconazole resistant C. albicans isolate and it was analyzed by a theoretical three-dimensional structure of the ERG11p.
ConclusionThe L321F mutation in C. albicans ERG11 gene may be associated with fluconazole resistance.
Durante años, el fluconazol se ha utilizado para tratar las infecciones por Candida. Sin embargo, el uso indiscriminado de este antimicótico ha favorecido la aparición de cepas resistentes. Se han descrito mutaciones en el gen ERG11 como uno de los principales mecanismos de resistencia en especies de Candida.
Objetivos En el presente estudio se investigaron las mutaciones de sentido erróneo en genes ERG11 de aislamientos de Candida albicans, glabrata y tropicalis previamente examinados mediante pruebas de sensibilidad a fluconazol.
MétodosLa detección de las mutaciones de este gen se realizó en 19 aislamientos clínicos de Candida (8C. albicans, 5C. glabrata y 6C. tropicalis) sensibles y resistentes a fluconazol. El gen se amplificó mediante reacción en cadena de la polimerasa (PCR) con cebadores específicos para cada especie de Candida y se analizaron mediante secuenciación automatizada.
ResultadosSe identificaron 14 mutaciones de sentido erróneo diferentes, 5 de las cuales no habían sido descritas previamente. Entre ellas, se identificó una nueva mutación L321F en un aislamiento de C. albicans resistente a fluconazol y que fue analizada mediante una estructura tridimensional teórica de ERG11p.
ConclusiónLa mutación L321F del gen ERG11 de C. albicans puede asociarse a resistencia a fluconazol.
Azoles are the most prescribed antifungal drugs in clinical practice, and the indiscriminate use of these drugs, especially fluconazole, has favored the emergence of resistance in Candida isolates. Different mechanisms of resistance to azole-based antifungal agents can occur simultaneously in resistant isolates. One of these mechanisms involves changes in the molecular configuration of the azole target enzyme 14α-demethylase (Erg11p) due to mutations in the encoding gene, ERG11, leading to a decrease in the affinity of the drug for the protein.7,14 Although more than 140 different missense mutations have been found in Candida albicans clinical isolates and documented in several reports,7,9,11,13,14,17,23 there is only one Latin American study, which was performed in Brazil and described 14 new mutations in the ERG11 gene of C. albicans clinical isolates from HIV-infected patients.7 Concerning non-C. albicans species, there are two studies on Candida tropicalis11,20 and four related to Candida glabrata.4,5,18,21 These reports documented only one missense mutation, an Y132F substitution in C. tropicalis.20 The aim of this study was to screen amino acid substitutions in Erg11p arising from missense mutations in the ERG11 genes of C. albicans, C. glabrata and C. tropicalis isolates.
Eight C. albicans, five C. glabrata and six C. tropicalis clinical isolates were evaluated in the study. They were obtained from different infected sites of 19 patients hospitalized in the city of São Paulo, Brazil (Table 1). The antifungal susceptibility to fluconazole was determined using the microdilution reference procedure of EUCAST for fermentative yeasts. The minimal inhibitory concentration values ≤4mg/L were considered as susceptible (S), 16–32mg/L as intermediate (I), and ≥64mg/L as resistant (R).6 The EUCAST guidelines has refrained from giving C. glabrata breakpoints for fluconazole.
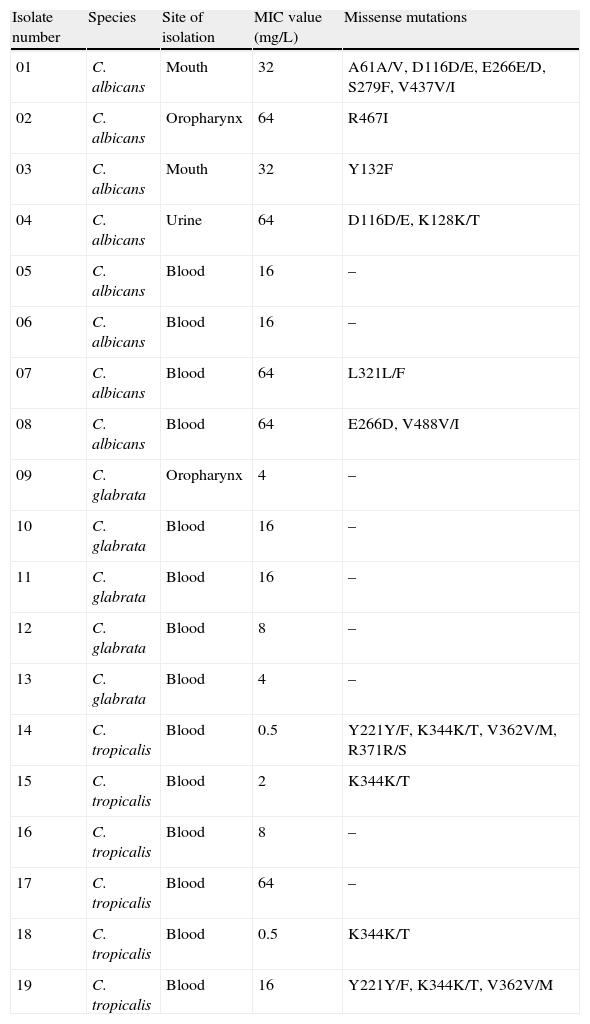
Site of isolation, fluconazole MIC values and missense mutations found in clinical isolates of C. albicans, C. glabrata and C. tropicalis evaluated in the study.
| Isolate number | Species | Site of isolation | MIC value (mg/L) | Missense mutations |
| 01 | C. albicans | Mouth | 32 | A61A/V, D116D/E, E266E/D, S279F, V437V/I |
| 02 | C. albicans | Oropharynx | 64 | R467I |
| 03 | C. albicans | Mouth | 32 | Y132F |
| 04 | C. albicans | Urine | 64 | D116D/E, K128K/T |
| 05 | C. albicans | Blood | 16 | – |
| 06 | C. albicans | Blood | 16 | – |
| 07 | C. albicans | Blood | 64 | L321L/F |
| 08 | C. albicans | Blood | 64 | E266D, V488V/I |
| 09 | C. glabrata | Oropharynx | 4 | – |
| 10 | C. glabrata | Blood | 16 | – |
| 11 | C. glabrata | Blood | 16 | – |
| 12 | C. glabrata | Blood | 8 | – |
| 13 | C. glabrata | Blood | 4 | – |
| 14 | C. tropicalis | Blood | 0.5 | Y221Y/F, K344K/T, V362V/M, R371R/S |
| 15 | C. tropicalis | Blood | 2 | K344K/T |
| 16 | C. tropicalis | Blood | 8 | – |
| 17 | C. tropicalis | Blood | 64 | – |
| 18 | C. tropicalis | Blood | 0.5 | K344K/T |
| 19 | C. tropicalis | Blood | 16 | Y221Y/F, K344K/T, V362V/M |
MIC – minimal inhibitory concentration.
If both alleles differ in sequence, a slash is used to indicate this heterozygosity.
Genomic DNA samples from all isolates were obtained employing the protocol of van Burik et al.19 with modifications. Candida species were confirmed by ITS amplification according to Luo et al. methodology12. The C. albicans ERG11 gene was amplified by PCR with previously described primers.23 Newly primers designed in this study were used to amplify C. glabrata and C. tropicalis ERG11 genes (CgP1F 5′-ACTACAATAACA TGTCCACTGA-3′ and CgP1R 5′-GGGTGGTCAAGTGGGAGTAA-3′; CgP2F 5′-AGCTGCTTACTCCC ACTTGACC-3′ and CgP2R 5′-AGCTTGTTGGGCATGGT CTCTC-3′; CgP3F 5′-GCCCAACAAGCTATCTCTGGTA-3′ and CgP3R 5′-TGT TTGGAATAGCGACA TCTCTC-3′; CgP4F 5′-CCAAACACTTCCTACGTTGTCC C-3′ and CgP4R 5′-GCATCTAGTACTTTTGTTCTGGATG-3′; CtP1F 5′-TCTTTT GTCAACACAGTA ATGGC-3′ and CtP1R 5′-AACACCTTTACCAAAAACAGGA G-3′; CtP2F 5′-CTCCTGTTTTTGGTAAAGGTGTT-3′ and CtP2R 5′-TGGATCAATATCACCGC TTTCTC-3′; CtP3F 5′-GCGGTGATATTGATCCAAAGAG-3′ and CtP3R 5′-TGGTATGAGCATAACCGGCAGA-3′; CtP4F 5′-TGCCGGTTATGCTCATACCA GT-3′ and CtP4R 5′-CTACTCCATGGTGATCTAAACC-3′). Amplification products were purified and sequenced using a MegaBACE-1000 analyzer (GE Healthcare, USA). PCR and sequencing were performed twice in both forward and reverse directions for all DNA samples to confirm the obtained data. The entire ERG11 open reading frame sequence from each isolate was aligned with previously described ERG11 sequences (GenBank accession: C. albicans X13296, C. glabrataDQ060157 and C. tropicalisAY942645) using ClustalW2.10 The similarity values between Candida reference strains and GenBank sequences were 99% for C. albicans and 100% for C. glabrata and C. tropicalis. The nucleotide changes found in the ERG11 sequences were characterized using the yeast differential genetic code for the CUN codon.15
A total of 21 missense mutations were found: 12 in C. albicans and nine in C. tropicalis isolates (Table 1). Seventeen of these mutations occurred in a single allele (heterozygous), and four were present in both alleles (homozygous). Among all the missense mutations, 14 different amino acid substitutions were identified, including five that have not been previously reported: L321F in C. albicans; and Y221F, K344T, V362M and R371S in C. tropicalis (Table 1). Although in this study the D116E, K128T, E266D, V437I and V488I mutations were identified only in C. albicans resistant isolates, they have been previously reported in both azole-susceptible and azole-resistant isolates, suggesting that these substitutions are probably not associated with resistance.7,13,14 Likewise, the novel mutations Y221F, K344T, V362M and R371S identified in C. tropicalis do not seem to be associated with fluconazole resistance once they were found in susceptible isolates. In contrast, four C. albicans mutations found in this study (A61V, Y132F, S279F and R467I) have been reported to be restricted to isolates with decreased susceptibility to fluconazole.7,14
No missense mutation was identified in C. glabrata isolates. This finding seems to be linked to the fact that other resistance mechanisms may be operating. Another hypothesis is that C. glabrata suffers less selective pressure in response to fluconazole because of its ability to collect human serum cholesterol and insert it into its plasma membrane replacing the ergosterol.1,16 On the other hand, the EUCAST guidelines indicate that a significant number of infections involving C. glabrata exhibit fluconazole MICs of 2–32mg/L without resistance mechanisms. Therefore, the C. glabrata isolates chosen in this study could in fact be fluconazole susceptible strains explaining why no mutations were found in ERG11 gene.
The novel mutation L321F was identified in a resistant C. albicans (isolate 7, Table 1) isolated from the bloodstream of a patient naïve for systemic antifungal treatment. However, he had been hospitalized and thus the nosocomial setting may have favored the emergence of a resistant yeast strain.17 Interestingly, the mutation L321F (A963C) was found in a single allele without association with any other mutation. This result suggests that heterozygosity is enough to generate a resistance phenotype and a forward mutation at the second allele or gene conversion may produce a higher level of resistance. We further sequenced and analyzed 24 C. albicans fluconazole-susceptible isolates (MIC values ranging from 0.25 to 2mg/L) to verify whether the L321F mutation was also present in low-MIC isolates. As a result, some known missense mutations were identified such as D116E, K128T, E266D, V437I and V488I. However, the new L321F mutation was not found in any of these isolates (data not shown) indicating that it might be linked only to fluconazole-resistant isolates.
The amino acid resulting from L321F mutation has different physical properties to the wild type; lysine is highly hydrophilic whereas phenylalanine is strongly hydrophobic. This change could possibly generate a modified secondary protein structure because the amino acid interactions with water are crucial to protein stability and folding.3 As described earlier, the molecular configuration of 14α-demethylase is critical for the fluconazole binding and this change may lead to a decrease in the affinity of the drug for the enzyme.7,14
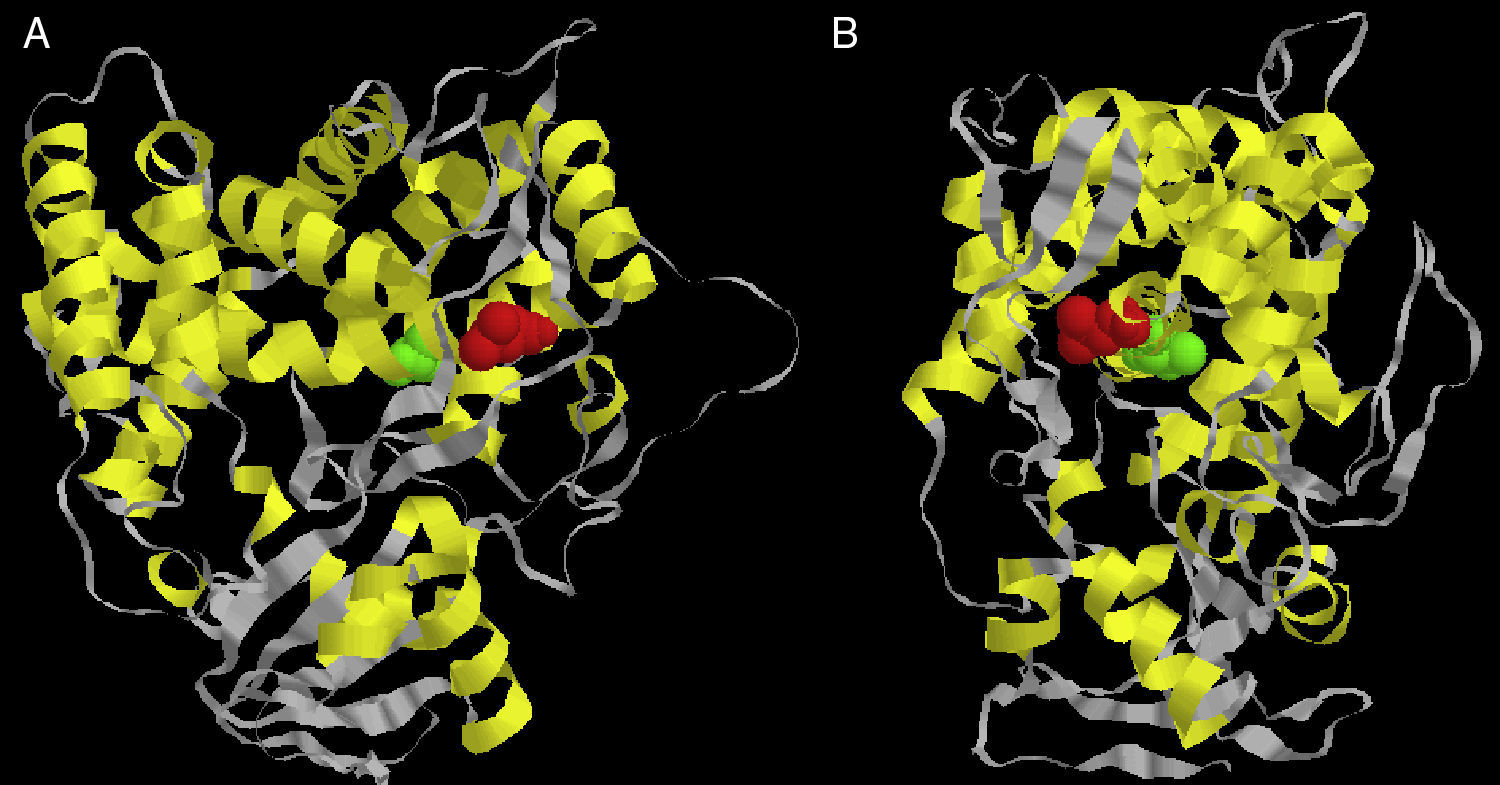
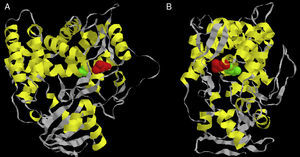
Considering there is no crystal structure of Erg11p available in literature, we designed a three-dimensional theoretical model based on the Erg11p sequence homology with the X-ray crystal structure of the 14α-demethylase from Mycobacterium tuberculosis. This model was obtained using 3D-JIGSAW software following the instructions provided by the developers.2 According to this model, the L321F mutation is located in the central helix of the molecule (Fig. 1A and B). The central helix is a region described as the enzyme binding site and crucial to fluconazole affinity.8,22 Therefore, this mutation may alter the three-dimensional structure of the CaERG11p resulting in a modified affinity to azoles.
Front (A) and side (B) views of the predicted three-dimensional structure of C. albicans Erg11p. The red spheres represent the L321F mutation found in the study, and the green spheres represent the T315A mutation evaluated in the Lamb et al.9 report.
Lamb et al.9 worked with a similar theoretical model and predicted that the T315A mutation (located at the central region of Saccharomyces cerevisiae Erg11p) was able to reduce the enzyme activity by 2-fold decreasing its affinity for azoles. In addition, transformant yeasts carrying T315A mutation showed 4-fold increased azole resistance. The substitution of threonine for alanine at residue 315 disturbed the protein structure, as indicated by the change from 448nm to 445nm in the Soret peak maximum. This altered conformation is probably responsible for the change in azole affinity.9 To date, this mutation has not been found in clinical isolates of C. albicans or other related species.9,14 However, when this T315A mutation was inserted into our theoretical model, it was possible to verify that it is located in the central helix of the enzyme near L321F mutation (Fig. 1A and B). The proximity between L321F and T315A mutations could indicate that the new L321F mutation might also be involved in the development of resistance to fluconazole. However, further experimental approaches such as L321F cloning into fluconazole-susceptible C. albicans strains and the evaluation of other resistance mechanisms are needed to confirm this association.
We thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support.