Organisms have evolved different strategies to respond to oxidative stress generated as a by-product of aerobic respiration and thus maintain the redox homeostasis within the cell. In particular, fungal pathogens are exposed to reactive oxygen species (ROS) when they interact with the phagocytic cells of the host which are the first line of defense against fungal infections. These pathogens have co-opted the enzymatic (catalases, superoxide dismutases (SODs), and peroxidases) and non-enzymatic (glutathione) mechanisms used to maintain the redox homeostasis within the cell, to resist oxidative stress and ensure survival within the host. Several virulence factors have been related to the response to oxidative stress in pathogenic fungi. The opportunistic fungal pathogen Candida glabrata (C. glabrata) is the second most common cause of candidiasis after Candida albicans (C. albicans). C. glabrata has a well defined oxidative stress response (OSR), which include both enzymatic and non-enzymatic mechanisms. C. glabrata OSR is controlled by the well-conserved transcription factors Yap1, Skn7, Msn2 and Msn4. In this review, we describe the OSR of C. glabrata, what is known about its core elements, its regulation and how C. glabrata interacts with the host.
This manuscript is part of the series of works presented at the “V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi” (Oaxaca, Mexico, 2012).
Los microorganismos han establecido diferentes estrategias para controlar el estrés oxidante generado durante la respiración aeróbica y, por consiguiente, mantener la homeostasia redox en la célula. En particular, los hongos patógenos se exponen a especies reactivas del oxígeno cuando interactúan con las células fagocíticas del huésped que son la primera línea de defensa contra estos agentes infecciosos. Estos patógenos han reclutado sistemas enzimáticos (catalasas, superóxido dismutasas y peroxidasas) y no enzimáticos (glutatión) que normalmente utilizan para mantener la homeostasis redox en la célula, para resistir frente al estrés oxidante y garantizar la supervivencia dentro del huésped. Varios factores de virulencia se han relacionado con la respuesta al estrés oxidante de los hongos patógenos. El hongo patógeno oportunista Candida glabrata (C. glabrata) es la segunda causa más frecuente de candidiasis después de Candida albicans (C. albicans). C. glabrata tiene una respuesta bien definida al estrés oxidante, que incluye sistemas enzimáticos y no enzimáticos y está regulada por los factores de transcripción Yap1, Skn7, Msn2 y Msn4. En esta revisión, describimos los elementos de la respuesta de C. glabrata a dicho estrés, cómo se regula y cómo C. glabrata interacciona con el huésped.
Este artículo forma parte de una serie de estudios presentados en el «V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi» (Oaxaca, México, 2012).
Yeast cells growing in an aerobic environment are exposed to reactive oxygen species (ROS) such as the superoxide anion (O2−), hydrogen peroxide (H2O2) and hydroxyl radical (OH). ROS are formed as by-products, during normal aerobic metabolism, and could damage all biomolecules and cause cell death.14 However, in order to maintain the redox homeostasis in the cell, a variety of enzymatic (catalases, SODs, and peroxidases) and non-enzymatic (glutathione) defense mechanisms are induced. This response is tightly controlled and is known as oxidative stress response (OSR).
Phagocytes are the first line of host defense against fungal infections. Upon interaction with the pathogen, phagocytes rapidly produce ROS through the NADPH oxidase complex.1 Deficiencies in the NADPH oxidase result in increased susceptibility to fungal and bacterial infections underlying the importance of the oxidative killing of microbes. These data show how the pathogens antioxidant systems are necessary to survive the oxidative stress generated by host phagocytes during infection.22 Until recently, it was not well-known how Candida glabrata responds to host cell phagocytosis and how it can survive and persist inside the phagolysosome.
C. glabrata is a haploid yeast found as a commensal in healthy individuals, but causes serious infections in immune compromised humans. In the past three decades, C. glabrata has become the second most common cause of candidiasis after Candida albicans (C. albicans). C. glabrata infections are difficult to treat and often result in high mortality in immune compromised hospitalized patients.2 In the past 10 years, the molecular mechanisms enabling C. glabrata to become a successful human pathogen have been described. C. glabrata OSR appears to play a central role in the survival inside macrophages: C. glabrata neutralizes the ROS generated by the macrophage38 by inducing antioxidant defenses,28,29 inhibits the maturation of the phagolysosome32 and ultimately replicates inside phagosomes.15 In this review, we will focus on the OSR of C. glabrata.
Enzymatic defensesCatalasesIn aerobic organisms, catalases are ubiquitous enzymes that decompose H2O2 to water and oxygen. They function as scavengers of H2O2 to maintain the redox balance in the cell. Given that H2O2 has been found to be a signaling molecule, catalases are also important for growth regulation and development.25C. glabrata has one catalase CgCta15 (Table 1), a mono-functional 57-kDa heme-containing protein, classified as a small subunit catalase and is 85% similar to the Saccharomyces cerevisiae peroxisomal catalase, ScCta1. Although CgCta1 does not have a canonical C-terminal peroxisomal targeting signal (PTS1),5 it has been shown to localize in the cytosol and accumulates in peroxisomes during respiration and inside phagocytic cells.28 Furthermore, the expression of CgCTA1 is induced in the presence of oxidative stress and in carbon source deprivation.29 Interestingly, the CgCTA1 locus does not maintain genic order when compared to other sequenced hemi-ascomycete yeasts.23 In particular, the upstream region of CTA1 is approximately 4.5kb, much longer than the average 454bp intergenic regions of C. glabrata.36
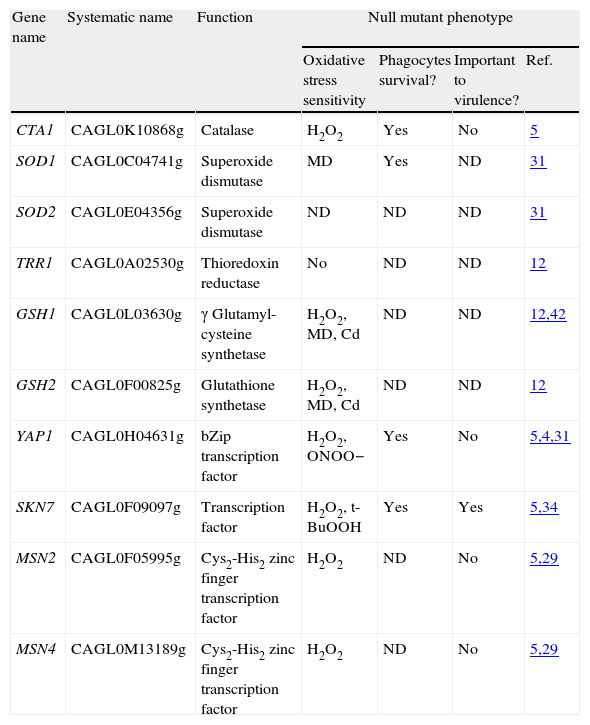
Oxidative stress response genes in Candida glabrata.
| Gene name | Systematic name | Function | Null mutant phenotype | |||
| Oxidative stress sensitivity | Phagocytes survival? | Important to virulence? | Ref. | |||
| CTA1 | CAGL0K10868g | Catalase | H2O2 | Yes | No | 5 |
| SOD1 | CAGL0C04741g | Superoxide dismutase | MD | Yes | ND | 31 |
| SOD2 | CAGL0E04356g | Superoxide dismutase | ND | ND | ND | 31 |
| TRR1 | CAGL0A02530g | Thioredoxin reductase | No | ND | ND | 12 |
| GSH1 | CAGL0L03630g | γ Glutamyl-cysteine synthetase | H2O2, MD, Cd | ND | ND | 12,42 |
| GSH2 | CAGL0F00825g | Glutathione synthetase | H2O2, MD, Cd | ND | ND | 12 |
| YAP1 | CAGL0H04631g | bZip transcription factor | H2O2, ONOO− | Yes | No | 5,4,31 |
| SKN7 | CAGL0F09097g | Transcription factor | H2O2, t-BuOOH | Yes | Yes | 5,34 |
| MSN2 | CAGL0F05995g | Cys2-His2 zinc finger transcription factor | H2O2 | ND | No | 5,29 |
| MSN4 | CAGL0M13189g | Cys2-His2 zinc finger transcription factor | H2O2 | ND | No | 5,29 |
Abbreviations: ND=not determined; H2O2=hydrogen peroxide; t-BuOOH=tert-butyl hydroperoxide; ONOO−=peroxynitrite; MD=menadione; Cd=cadmium; bZIP=basic leucine zipper transcription factor.
The in vitro resistance of C. glabrata to H2O2 is very high, compared to S. cerevisiae or C. albicans5 and this in vitro resistance is mediated by the CgCta1; however, in a murine model of systemic infection the cta1▵ mutant was not affected in the colonization phenotype of target organs in vivo.5 This suggested that other antioxidant molecules could be compensating for the absence of CgCta1. Analysis of the double mutants lacking glutathione or thioredoxin suggested that theses antioxidant molecules are not responsible for compensating in vivo the absence of CgCta1.10 To date, the role of CgCta1 as a virulence factor has not been established as in other pathogenic fungi.8
Superoxide dismutasesSODs are metalloenzymes that catalyze the dismutation of O2− into H2O2 and oxygen. These enzymes protect cells against oxidative damage and scavenge O2− radicals generated during aerobic metabolism. Based on their metal cofactor, SODs are classified in three families: copper/zinc (Cu,ZnSOD), iron or manganese SODs (FeSOD and MnSOD) and nickel SODs (NiSOD).7 Most eukaryotes contain two SODs, a MnSOD localized in the mitochondrial matrix37 and a highly abundant Cu,ZnSOD present in cytosol and mitochondrial inter-membrane space.26C. glabrata has two SOD genes, CgSOD1 (Cu,ZnSOD) and CgSOD2 (MnSOD) (Table 1). Interestingly, it has been shown that Cryptococcus neoformans, C. albicans and Histoplasma capsulatum SODs are required for survival against macrophages13,24,40; however, the survival rate of C. glabrata sod1Δ mutant in macrophages was not diminished.29
In S. cerevisiae, Schizosaccharomyces pombe and C. albicans, SOD1 expression is induced by oxidative stress,6,18 and regulated by Yap1, a key transcriptional regulator of the OSR.16,17,42 In contrast, C. glabrata CgSOD1 and CgSOD2 are constitutively expressed even in the presence of oxidative stress and independent of CgYap1.29 CgSOD1 and CgSOD2 expression are highly induced under glucose starvation. Interestingly, the survival rate of C. glabrata sod1Δ mutant in macrophages was diminished only in combination with a mutation in CgYap1.29
Glutathione and thioredoxinRedox homeostasis is necessary to support important cellular processes. The oxidation status of thiols is maintained and rapidly restored by the action of two redox-balancing systems: glutathione (GSH) and thioredoxin pathways.12 In yeast, the GSH system is constituted by GSH, glutaredoxins and a glutathione reductase. Similarly, the cytoplasmic thioredoxin pathway comprises two thioredoxins and a thioredoxin reductase.35C. glabrata has the majority of the components of both systems (Table 1); however, their role in the OSR of C. glabrata has recently been described.10,39
GSH is an essential tripeptide composed of glycine, cysteine and glutamate synthesized by the sequential action of Gsh1 and Gsh2.21 Through its thiol group, GSH reacts directly with reactive species or acts as cofactor with specific enzymes (glutaredoxins, glutathione peroxidases or glutathione transferases) to detoxify ROS or xenobiotics. In C. glabrata, GSH1 is essential39; however, a suppressor mutation in CgPRO2 was isolated in the absence of GSH1. The pro2-4 suppressor mutation could be acting through the synthesis of residual amounts of GSH.10 Additionally, C. glabrata cells lacking glutathione are sensitive to oxidative stress and have a reduced late chronological life span.10 Surprisingly, the S. cerevisiae specific GSH transporter (ScOPT1) is not present in C. glabrata; however, there is evidence suggesting external uptake of GSH by an unknown transporter.10 The differences between S. cerevisiae and C. glabrata in the GSH pathway and the absence of the GSH specific transporter suggest that GSH may play an important role during pathogenesis.
The thioredoxin system is a pivotal player in the redox homeostasis since some components affect the oxidative stress resistance in many microorganisms. In C. glabrata, there is evidence that suggest that the cytoplasmic thioredoxin system plays a minor role in H2O2 adaptation; however, the inability to disrupt simultaneously the three systems (GSH, thioredoxin and catalase) in C. glabrata suggests that these pathways could be complementing each other in order to respond to oxidative stress.10
Metallothioneins, phytochelatins and pigmentsSome metals are essential micronutrients for physiological processes. Copper, zinc and other non-essential heavy metal ions, such as cadmium, lead, and mercury, are highly reactive and can produce oxidative stress. To diminish the toxic effects of heavy metals, some yeast synthesize phytochelatins (PCs), which are enzymatically synthesized cysteine-rich peptides derived from GSH and metallothioneins (MT), oligomeric thiolated molecules that act as chelators.11 In the C. glabrata genome, two MT isoforms have been identified: MT-I (CAGL0D01265g) and MT-II (CAGL0H04257g).19C. glabrata appears to chelate cadmium with phytochelatin-like molecules20; however, the phytochelatin synthase coding gene has not been identified.
Pigments are important factors in pathogenic fungi for survival in their host. Production of the dark pigment melanin has been linked to pathogenicity. Recently it was shown that C. glabrata produces a pigment that protects against H2O2 and the attack by human neutrophils. This pigment is a by-product of the Ehrlich pathway of tryptophan degradation and its production is mainly driven by the aromatic aminotransferase I (Aro8).3
Transcriptional regulation by reactive oxygen speciesCgYap1A number of well-conserved transcription factors have been identified that control the OSR in C. glabrata.5,28,29 Yap1 is a bZip transcription factor that contains cysteine rich domains in its N- and C-terminal portions. The transcription factor Yap1 of C. glabrata controls the OSR and accumulates transiently in the nucleus during phagocytosis.28,29 In a genome wide analysis in C. glabrata, one set of genes (CTA1, TRR1/2, TSA1/2, TRX2, GPX2 and CCP1) were dependent on the presence of both, Skn7 and Yap1, and defined the core of the OSR. A second Yap1-regulated group included genes encoding proteins with aldo-ketoreductase and oxidoreductase activities (ADH6, GRE2, SCS7 and OYE2).29 Interestingly, the loss of CgYap1 had no impact on virulence in both primary mouse macrophages or in a murine model of systemic infection.4,29 It is possible that either the genes controlled by CgYap1 are dispensable to survive within the host or there are redundant mechanisms that compensate the lack of the genes controlled by this transcription factor.
CgSkn7Skn7 is an oxidative and cell wall stress-responsive transcription factor highly conserved among fungi. It has been shown that in C. glabrata Skn7 controls the expression of a set of genes including TRX2, TRR1, TSA1 and CTA1 in response to the presence of H2O2 and is required for the adaptation response to oxidative stress.5,30,34 Skn7 has been shown to be important for virulence in a murine model of systemic infection31; however, Skn7 was dispensable for prolonged survival in macrophages.29
CgMsn2 and CgMsn4The Cys2-His2 zinc finger transcription factors Msn2 and Msn4 mediate the general stress response and functions in parallel with Yap1 and Skn7 to mediate the OSR in C. glabrata.5,27 However, the function of Msn2 and Msn4-like proteins has diverged significantly among fungi. In a Drosophila melanogaster infection model, it was shown that Msn2 does not have a major impact on virulence.27 Genes functionally connected to stress response, such as HSP12, HSP42, DDR48, GPH1, GDB, TPS1, TPS2 or PGM2 displayed a CgMsn2 and Msn4-associated upregulation.27,41
Concluding remarksIn the past 10 years, some of the C. glabrata virulence factors that contribute to pathogenesis have been identified, and in particular specific efforts have been made to study the interplay between C. glabrata and the host. As a successful fungal pathogen, C. glabrata quickly detects and responds to metabolic changes and to oxidative stress through the induction of protective enzymes against oxidative stress (catalase and SODs) and non-enzymatic defense systems (GSH) (Fig. 1). The transcription factors, Msn2, Msn4, Skn7 and Yap1, play a central role in the regulation of OSR. The fact that these transcriptional factors are dispensable for virulence (except for Skn7) suggests that C. glabrata survival depends on redundant pathways that can compensate each other (Fig. 1). It is possible that the ability of C. glabrata to suppress the production of ROS ensures its survival during phagocytosis.33 Interestingly, although the structure of the MAP kinase pathways is relatively conserved among fungi, the role of MAPK pathways in C. glabrata remains largely unexplored despite the fact that these pathways are important for virulence.9 The identification and characterization of the proteins involved in the detection of the oxidative signal will be essential to understand how C. glabrata evades killing by the phagocytic cells. Our knowledge about the OSR of C. glabrata is increasing rapidly and, in the next few years, future research will reveal major new insights into the regulatory networks and signal transduction pathways that regulate and coordinate the stress regulons in C. glabrata.
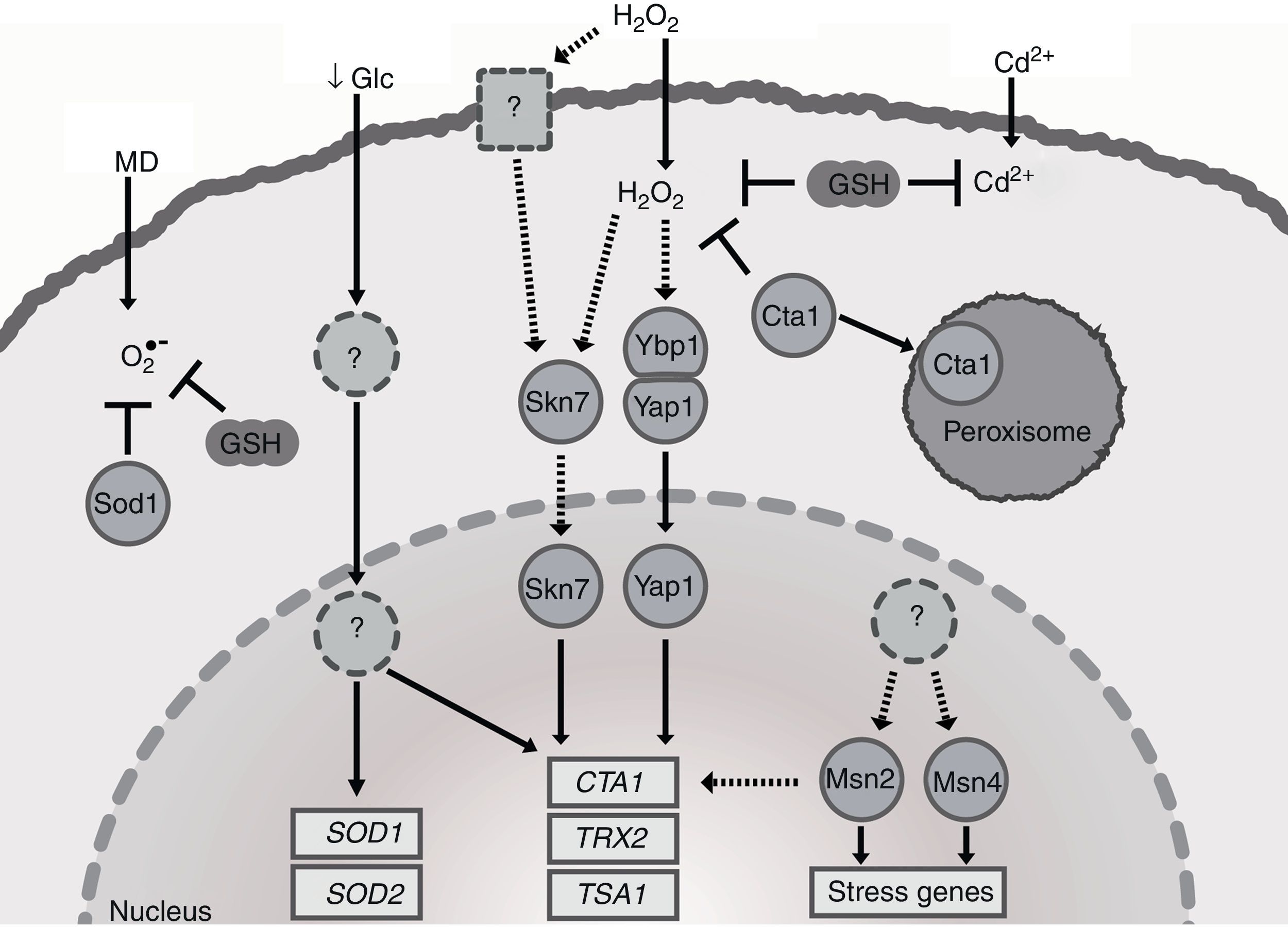
Pathways of the OSR in Candida glabrata. C. glabrata responds with antioxidant defenses against external sources of oxidative stress such as menadione (MD), hydrogen peroxide (H2O2) and cadmium (Cd2+). The peroxide-induced signal transduction pathway of C. glabrata is still unknown, but data suggest that H2O2 activates the transcription factors Yap1, Skn7 and Msn4. Yap1 response to H2O2 requires Ybp1. Yap1 and Skn7 activate transcription of the catalase (CTA1), thioredoxin (TRX2), and thioredoxin peroxidase (TSA1). The catalase detoxifies H2O2 in cytoplasm and peroxisomes. To neutralize the superoxide (O2−), C. glabrata induces the superoxide dismutase (Sod1) and the synthesis of glutathione (GSH). Sod1 converts O2− to H2O2. GSH participates in maintaining the redox balance. The transcription of CTA1, SOD1 and SOD2 is induced by glucose starvation (↓Glc). SOD1 and SOD2 do not respond to H2O2. Msn2 and Msn4 are involved in the activation of the general stress response. The discontinuous arrows and dashed-line circles represent not-well established pathways.
The authors declare that they have no conflict of interests.
This work was funded by a Consejo Nacional de Ciencia y Tecnología (CONACYT) fellowship to M.B.M.C. (209276), E.O.Z. (233455), J.J.C. (48549), G.G.E. (48580), and I.C.V. (224300). This work was funded by a CONACYT grant No. CB-2010-01-153929 to A.D.L.P. and grant No. CB-2010-01-151517 to I.C.N.