Rhinosporidum seeberi is the etiologic agent of rhinosporidiosis, a disease of mucous membranes and infrequent of the skin and other tissues of humans and animals. Because it resists culture, for more than 100 years true taxonomic identity of R. seeberi has been controversial. Three hypotheses in a long list of related views have been recently introduced: 1) a prokaryote cyanobacterium in the genus Microcystis is the etiologic agent of rhinosporidiosis, 2) R. seeberi is a eukaryote pathogen in the Mesomycetozoa and 3) R. seeberi is a fungus. The reviewed literature on the electron microscopic, the histopathological and more recently the data from several molecular studies strongly support the view that R. seeberi is a eukaryote pathogen, but not a fungus. The suggested morphological resemblance of R. seeberi with the genera Microcystis (bacteria), Synchytrium and Colletotrichum (fungi) by different teams is merely hypothetical and lacked the scientific rigor needed to validate the proposed systems. A fundamental aspect against the prokaryote theory is the presence of nuclei reported by numerous authors and updated in this review. Moreover, Microcystis's and Synchytrium's ultra-structural and key cell cycle traits cannot be found in R. seeberi parasitic phase. The PCR amplification of a cyanobacteria 16S rDNA sequence from cases of rhinosporidiosis, while intriguing, will be viewed here as an anomaly due to contamination with environmental Microcystis or perhaps as an endosymbiotic acquisition of plastids from cyanobacteria ancestors. Thus, even if R. seeberi possesses prokaryote DNA, this does not prove that R. seeberi is a cyanobacterium. The placement of R. seeberi within the fungi is scientifically untenable. The isolation and the DNA analysis performed in a fungal strain, and the lack of appropriate controls are the main problems of this claim. Further studies are needed to validate R. seeberi's acquisition of prokaryote plastids and other issues that still need careful scrutiny.
Rhinosporidum seeberi es el agente etiológico de la rinosporidiosis, una enfermedad de las membranas mucosas y, con menos frecuencia, de la piel y otros tejidos. Debido a que se resiste a crecer en los medios de cultivo desde hace más de 100 años, la identidad taxonómica de R. seeberi ha sido motivo de controversia. Tres nuevas hipótesis en una larga lista de puntos de vista similares han sido introducidas: 1) la cianobacteria Microcystis es el agente etiológico de la rinosporidiosis, 2) R. seeberi es un patógeno eucariota en los Mesomycetozoa, y 3) R. seeberi es un hongo. La literatura revisada sobre los estudios realizados con microscopia electrónica, los datos histopatológico y, más recientemente, los datos de varios estudios moleculares, apoyan fuertemente la idea de que R. seeberi es un patógeno eucariota, pero no un hongo. La semejanza morfológica propuesta por algunos de que R. seeberi es similar a los miembros de los géneros Microcystis (bacteria), Synchytrium y Colletotrichum (hongos) es meramente hipotética y no tiene el rigor científico necesario para validar el sistema propuesto. Un aspecto fundamental en contra de la teoría procariota es la presencia de núcleos descrita por numerosos autores y que actualizamos en esta revisión. Además, las características ultra-estructurales de los géneros Microcystis y Synchytrium y de sus ciclos celulares no han sido encontradas en la fase parasitaria de R. seeberi. La amplificación por PCR de una secuencia del rADN 16S típica de las cianobacterias en muestras de casos de rinosporidiosis, aunque interesante, será considerada en esta revisión como una anomalía debido a la contaminación con el medio ambiente (Microcystis) o tal vez como una adquisición endosimbiótica de plastidios a partir de cianobacterias ancestrales. Así pues, aunque R. seeberi podría poseer ADN procariota, esto no demuestra necesariamente que R. seeberi sea una cianobacteria. La clasificación de R. seeberi dentro de los hongos es insostenible. El aislamiento de un hongo, los análisis de ADN realizados, y la ausencia de controles apropiados son los problemas más importantes de esta teoría. Más estudios serán necesarios para validar la adquisición de plastidios procariotas en R. seeberi, y otros temas que requieren un cuidadoso escrutinio.
Rhinosporidium seeberi is the causative agent of rhinosporidiosis in humans and animals.14,59,100 The disease affects mucous membranes and rarely the skin and/or the internal tissues of its infected hosts.14,60 The first report of rhinosporidiosis was published by Seeber88 and occurred in a 19-year-old Argentinean patient with breathing difficulties caused by a polyp that obstructed his nasal passages. In his thesis Seeber88 noted also that in 1892 Malbram, in Buenos Aires, Argentina, was the first to diagnose the disease in humans, but he did not publish his finding. Although the disease has been frequently diagnosed in the Americas and other areas of world, rhinosporidiosis is more prevalent in India and Sri Lanka than in any other geographic locations.14,100 The original published cases in Argentina, and many others recorded thereafter, established that R. seeberi develops in the infected hosts 10 to >450μm in diameter spherical structures (sporangia), some containing hundreds of endoconidia (sporoblast, trophocyte), as well as immature forms of different sizes (5–80μm) without endoconidia. Despite the large numbers of spherical structures in the infected tissues, it was soon apparent that R. seeberi resists culture and that a rhinosporidiosis infection cannot be induced in experimental animals.14,15 These unique features steamed a great controversy that still continues to the present days.9,35,42
As has been the case with a number of uncultivated pathogens, such as Lacazia loboi61,102 and Mycobacterium leprae,66,78,81 numerous investigators have claimed that they had successfully produced R. seeberi in culture.27,58,65 In general most of the isolated strains were later found to be common contaminants.34,66,103 A good example of this trend is the most recent report claiming that R. seeberi is not a mesomycetozoa or a bacterium, but a fungus (97). This new proposal was based on fungal strains recovered from cases of rhinosporidiosis and from a frozen clinical sample collected ten years ago from a patient with rhinosporidiosis in India (97, 98). Thus, strains obtained from well known uncultivated pathogens have always been suspect and their identities are subjected to endless controversies. However, with the advent of molecular tools most of these debates came to an end, although not for R. seeberi.9,25,42,69,103
Currently there are three main hypotheses on the taxonomic and phylogenetic positions of R. seeberi in the tree of life; the first argues that R. seeberi is a prokaryote cyanobacterium in the genus Microcystis,6,9 the second one claims that R. seeberi is a eukaryote microbe closely related to spherical fish pathogens in the Mesomycetozoa,35,42 and the most recent hypothesis claims that R. seeberi is a fungus. These conflicting views on the taxonomy and phylogeny of R. seeberi will be critically addressed in this review. To put into perspective these hypotheses, we will first review results from traditional studies of R. seeberi to define its main characteristics and its taxonomy by focusing on the histological and electron microscopy (EM) data that have been accumulated over the last 110 years. We will also provide a historical description of the current R. seeberi theories to finally critically address these positions.
Traditional nomenclature, systematic and current views on R. seeberiThe taxonomic classification of R. seeberi has been always contentious. Based on its parasitic spherical morphological features with the development of endoconidia, Seeber88 believed that it was a protist closely related to a Coccidium (Coccidium seeberia, Wernecki).16,20 Sebeer's former adviser Wernicke20 thought it could also be a fungus similar to Coccidium immitis, and Minchin and Fantham72 grouped the pathogen with the Haplosporidia. Battie19 classified it with the Neurosporidia, and Ashworth16 treated it as a fungus in the zygomycetes or possibly a chytridiomycete. Based on ultrastructural characteristics, Vanbreuseghem101 suggested that R. seeberi may have a relationship with the algae, whereas Laveran and Petit63 were the first to note the resemblance of Ichthyosporidum (currently Icthyophonus a pathogen of trout) with R. seeberi. Later Acevedo2 and Carini22 mentioned that R. seeberi also shared morphological attributes with Dermosporidium hylarum (Amphibiocystidium ranae).82 Interestingly, these two pathogens of fishes and frogs are currently classified with members of the Dermocystida in the Mesomycetozoa (see below).3,68
When Seeber88 recorded the first case of rhinosporidiosis, he carefully described R. seeberi's morphological features in infected tissues, but did not name the organism. Instead the resemblance of this pathogen with C. immitis, the etiologic agent of coccidioidomycosis,105 was mentioned. It is believed that Seeber's professor Robert Wernicke named the organism C. seeberia, as appeared in a small note on the “Tratado de Parasitología Animal” in 1903.20 Apparently, Wernicke suggested also the name “Coccidium seeberi” in an ephemeral publication in 1900 at the University of Buenos Aires, Argentina under the name “Programa de Zoología Médica”.16,32 Even though the 1900 original edition was no longer available, Ashworth16 acquired a copy of the document dated 1907 and stated that the species C. seeberi was validly introduced. Unaware of these proposals, Minchin and Fantham72 studied histological samples of an Indian rhinosporidiosis case, described earlier by O’Kinealy,77 and introduced the binomial Rhinosporidium kinealyi. Soon, other cases of rhinosporidiosis were also attributed to R. kinealyi, including the first case of the disease in the United Sates.19,107 In response to Minchin and Fantham's72 proposal, Seeber89 adopted the genus Rhinosporidium, but argues for the species priority of the original name.20 He suggested that the combination R. seeberi should be accepted. In 1913 Zschokke110 introduced yet another species Rhinosporidium equi after studying histological sections obtained from a horse with a nasal polyp in South Africa. However, in 1923 Ashworth16 revised all proposals and concluded that R. seeberi has priority over new proposed species. Thus, the species Rhinosporidium ayyari,11Rhinosporidium hylarum,2,22 and Rhinosporidium amazonicum1 introduced later became synonyms of R. seeberi.
Bizarre hypotheses claiming that the spherical structures found in rhinosporidiosis were self-assembled material made of a mixture of plant and human material or spheres of cellular waste due to the ingestion of tapioca, have been suggested.7,10,17 However, these extreme views were soon abandoned. Subsequently Alhuwalia et al.9 proposed a theory linking R. seeberi with a prokaryote cyanobacterium in the genus Microcystis, a view that challenged traditional studies placing R. seeberi among the eukaryote microbes.14 Concurrently, Herr et al.,42 and Fredricks et al.,35 based on morphological and the PCR amplification of the 18S small subunit ribosomal DNA sequences, concluded that this pathogen was an eukaryote microbe. Herr et al.,42 introduced the name Mesomycetozoa (between animals and fungi) to place R. seeberi with microbes displaying spherical phenotypes with endoconidia, such as Dermocystidium and Sphaerothecum destruens (the rosette agent). Based on molecular phylogenetic analysis methods and the available morphological data, the possibility of several host specific strains in the genus Rhinosporidium was also suggested.91 Thankamani99 and Thankamani and Lipin-Dev97 introduced the most recent hypothesis on the taxonomic placement of this unique pathogen. They claimed the isolation of a fungus from cases of rhinosporidiosis and thus, they believe this fungal strain must be the etiologic agent of rhinosporidiosis.
Historical studies on the parasitic life cycle of R. seeberiSeeber88 was the first to address R. seeberi's complex life cycle in the tissue of its infected hosts. His observations were later confirmed by numerous investigators studying the morphological characteristics of R. seeberi in humans and animals with rhinosporidiosis.14 After Seeber's thesis,88 several comprehensive studies on the morphological and life cycle features of this unique pathogen were published by Acevedo,2 Ahsworth,16 Grover,39 Kurunaratne,59 Melo,67 and Thianprasit and Thagerngpol.100
Because R. seeberi resists culture, its true ecological niche is still unknown.16,26,59 Based on the proximity of the affected patients to wet environments, most investigators agree with the concept that R. seeberi must be located in aquatic niches. Although the occurrence of rhinosporidiosis in dry areas of the Middle East seems to contradict this view, it is quite possible that R. seeberi had evolved also to develop resistant structures in dry environments.14 Despite the finding of rhinosporidiosis cases in arid areas, it is widely accepted that the infecting units (possibly non-flagellate spores) are more likely located in aquatic environments and may contact affected hosts through traumatic lesions, leading to the implantation of the pathogen in the host's tissues (see below).26 Once in the host, the infecting unit of unknown origin increases in size (10 to >450μm in diameter) and through cytoplasmic cleavage produces hundreds of endoconidia within the spherical cells, which subsequently are released through a single pore and reinitiate a life cycle.47,54
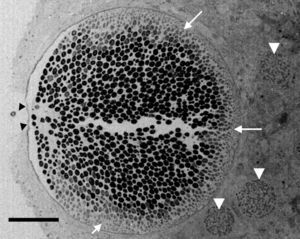
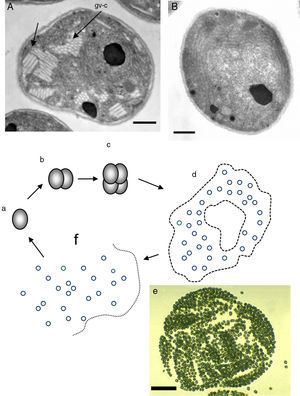
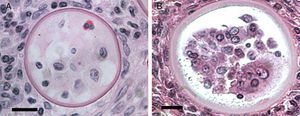
According to numerous studies using stained tissue sections and electron microscopy (EM),2,18,39,45,48,53,75,95,96R. seeberi's life cycle starts with the release of mature endoconidia through a small pore in the cell wall of the mature sporangium (Fig. 1, this review). A film depicting R. seeberi endoconidia release through a pore upon activation with water is available at: http://www.bld.msu.edu/Rhino. The diameter of an endoconidia ranges from 4 to 10μm. Numerous EM studies have also revealed that the endoconidia possess a well defined thin bilaminated cell wall (1–3μm) containing several small vesicles termed electron dense bodies (EDBs), a nucleus with a distinctive nucleolus, and a granular mucilaginous capsule, as those also shown in Fig. 2A, this review.2,16,47,54,59,75,82,87,95,101 Some investigators have further postulated that the EDBs within the endoconidium are the true generative infective units of R. seeberi and even have proposed the term “spore-morula” for the way these structures give origin to endosporulated sporangia.12,19,60 However, this theory has few followers and so far has not been fully accepted. Although some authors have reported the presence of nuclei and mitochondria in the endoconidia, these two organelles are extremely difficult to visualize either before or after the release of the endoconidia (Fig. 2A, this review). This is probably due to problems during the tissue fixative process prior to staining protocols.16,40,45,59,87,95,101,106
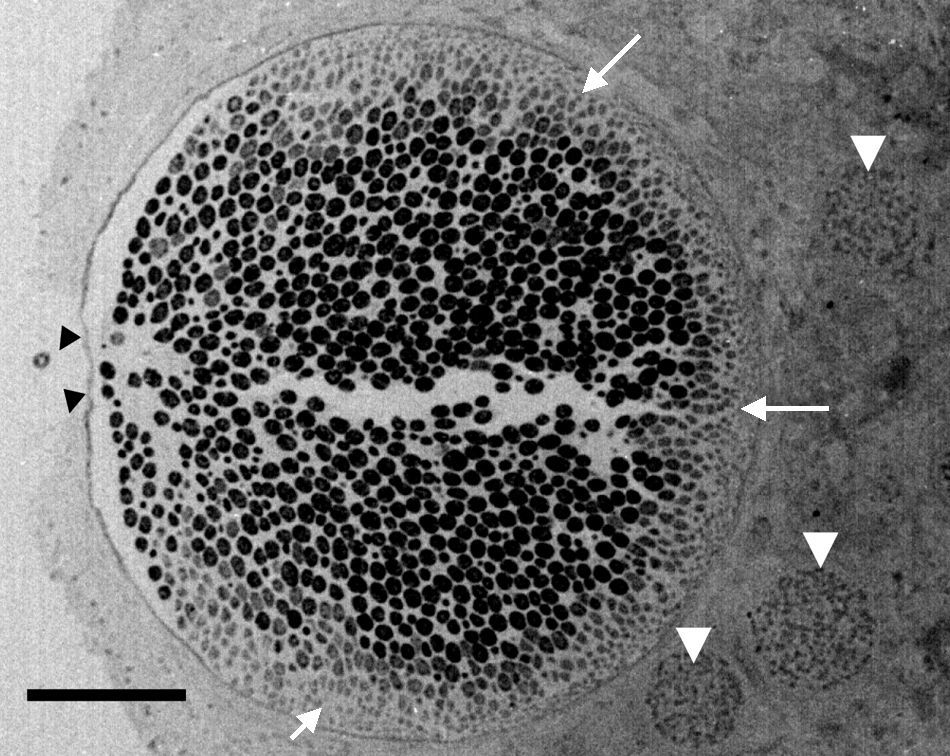
Electron microscopy tissue section showing a Rhinosporidium seeberi mature sporangium containing hundreds of endoconidia, one already outside the sporangium and the others in the process of being expelled through a pore (arrow heads) (a video depicting the endoconidia release is available at: http://www.bld.msu.edu/Rhino). Note the presence of a thin cell wall and the formation of three prominent inner layers (a clear space between the mature endoconidia and the cell wall) primarily located near the pore. At this magnification this structure appears as a single inner layer, but it comprises three well defined inner layers.40 The presence of fully developed endoconidia is observed at the center and toward the pore, whereas immature small usually oval endoconidia are found at the opposite site (white heads), a distinctive feature of mature sporangia. Three immature sporangia are also noted in the lower section of the mature sporangium (white arrow heads) (Bar=100(μm).
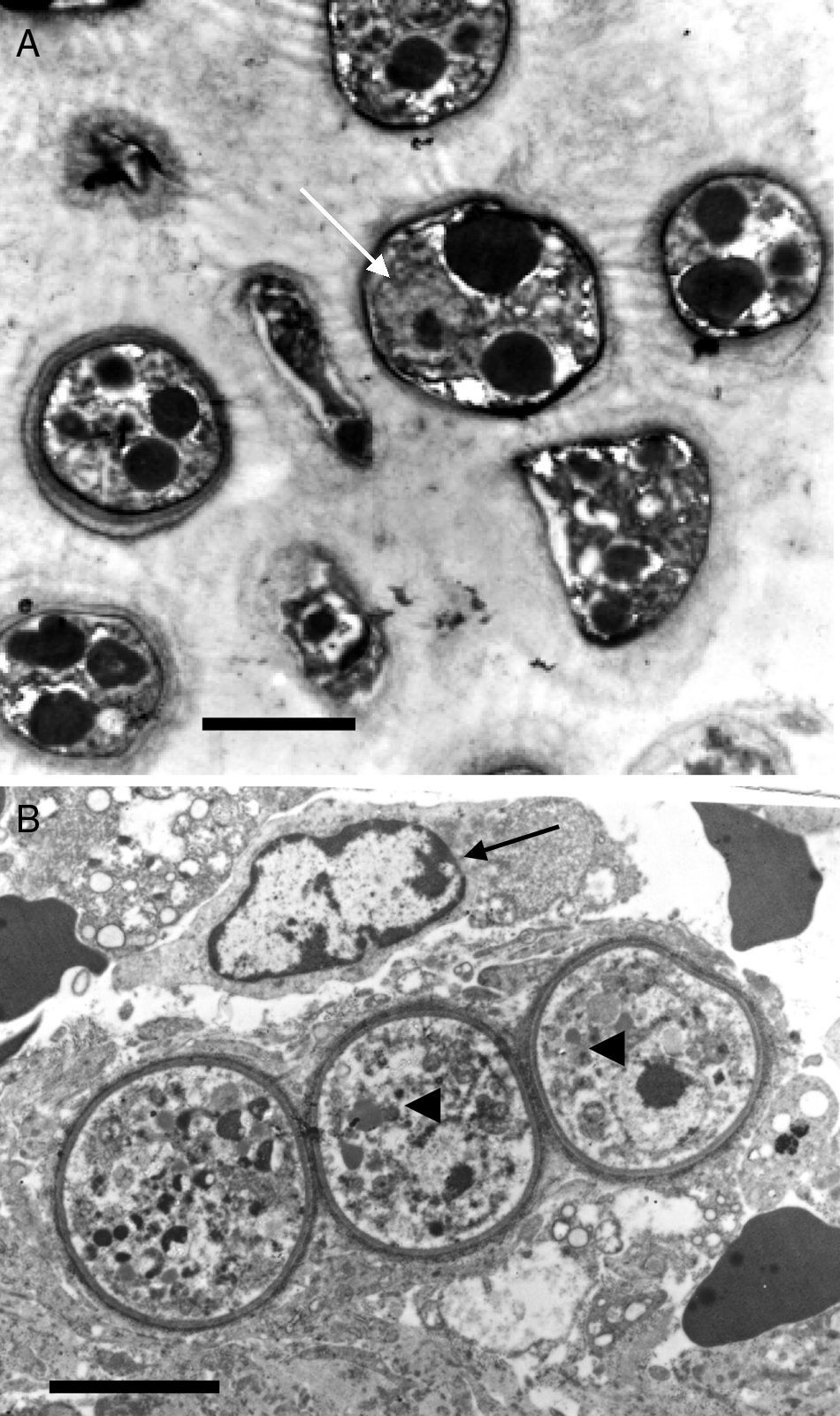
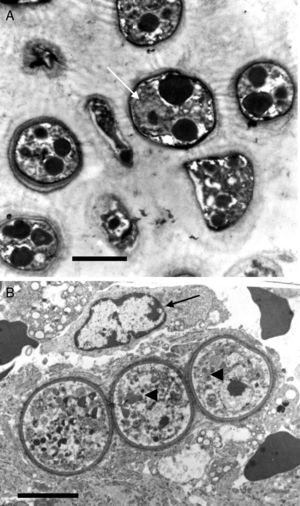
This electron micrograph shows several Rhinosporidium seeberi's purified endoconidia, one of them containing a nucleus with a prominent nucleolus (arrow) (Panel A). The endoconidia are characterized by a thin cell wall, numerous electron dense bodies (EDBs, dark vesicles), and the presence of a granular mucilaginous capsule (Bar=5(μm). Panel B shows three early juvenile sporangia (JS) within the host infected tissues, before the first nuclear division (Bar=15(μm). The presence of nuclei with prominent nucleolus is observed in two of the JS (arrow heads), which contrast with the nucleus displayed by the host's inflammatory cells (arrow). The JS on the left lacks nucleus because it was sectioned in a segment of the cell where the nucleus was not near. The presence of prominent cell wall and several vesicles typical of this stage is also noted.
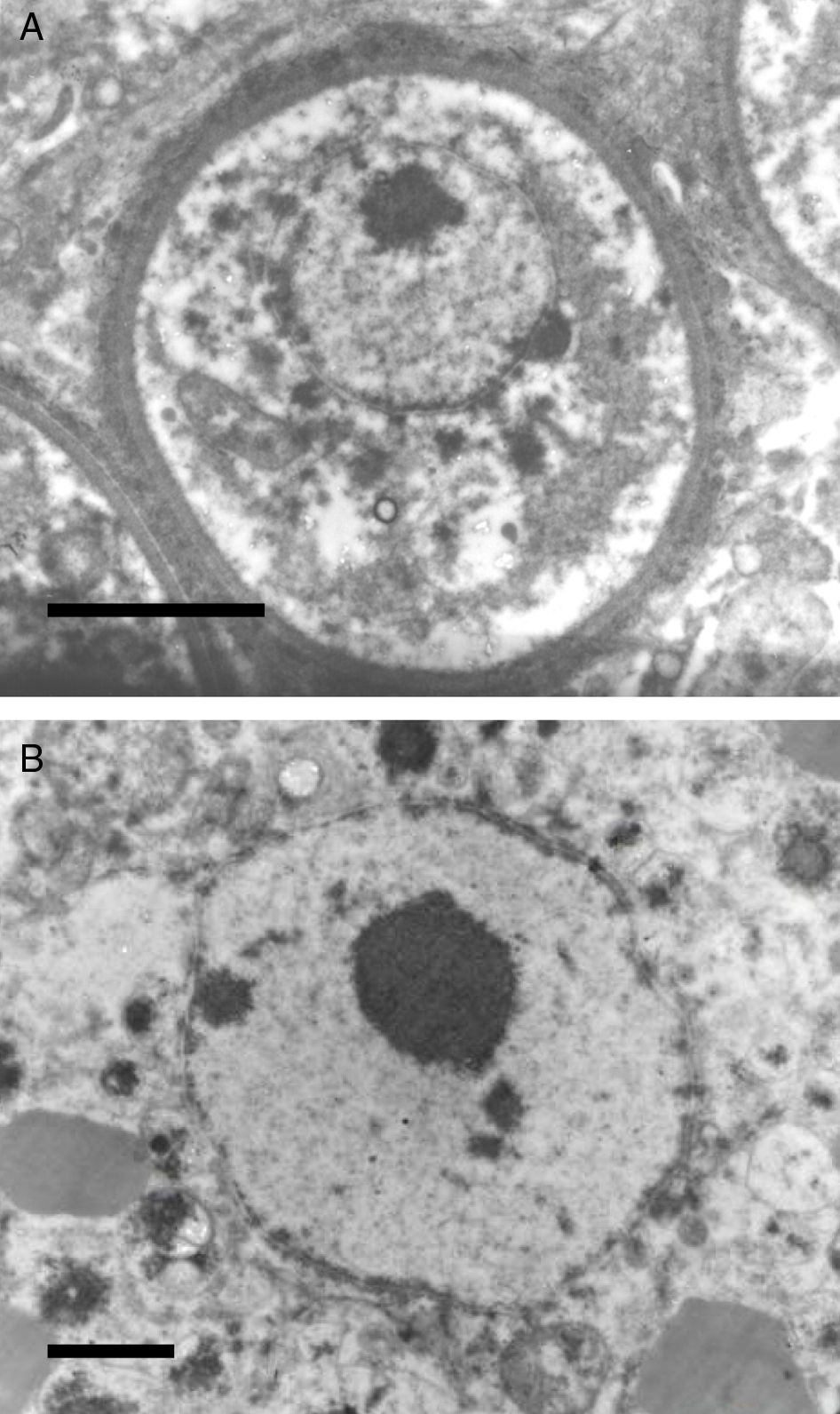
It has been postulated that after the release, the endoconidia increase in size (∼10–70μm) and by absorption loses all vesicles and EDBs to become a juvenile sporangium (JS) (Fig. 2B, this review).16,47,54,59,87 Based on EM studies, this stage is characterized by a prominent cell wall, a granular cytoplasm, the presence of a single central nucleus with a noticeable nucleolus enclosed by nuclear membrane, and some mitochondria (Figs. 2B and 3A, this review).2,16,46,54,59,75,87,101 The presence of endoplasmic reticulum, lipidic globules, and vacuolate structures has also been described.40,45,54,87 The juvenile sporangium increases in size and becomes an intermediate sporangium (IS) reaching ∼70–150μm in diameter (Fig. 3B, this review). Several investigators, including Acevedo,2 Ahsworth,16 Herr et al.,40 Kurunaratne,59 and others have reported at this stage the presence of a thick cell wall containing chitin, granular cytoplasm with numerous nuclei dispersed within the cytoplasm, numerous flat cristae mitochondria, the presence of an early pore, and one or more electron dense concentric rings termed “laminated bodies”. The latter structures have been considered by some as artifacts due to the tissue fixative process,67 or as structures made of chromatin by others.42,87 The presence of immature endoconidia at this stage is a rare finding. By nuclear cleavage the last nuclear division takes place. This event is followed by the development of a thin cell wall around each nucleus, giving rise to the formation of immature endoconidia, and thus the whole structure becomes an early mature sporangium.17,18,45,47 Most of the morphological features of mature sporangia such as the presence of inner layers and a visible pore are missing at this early stage.
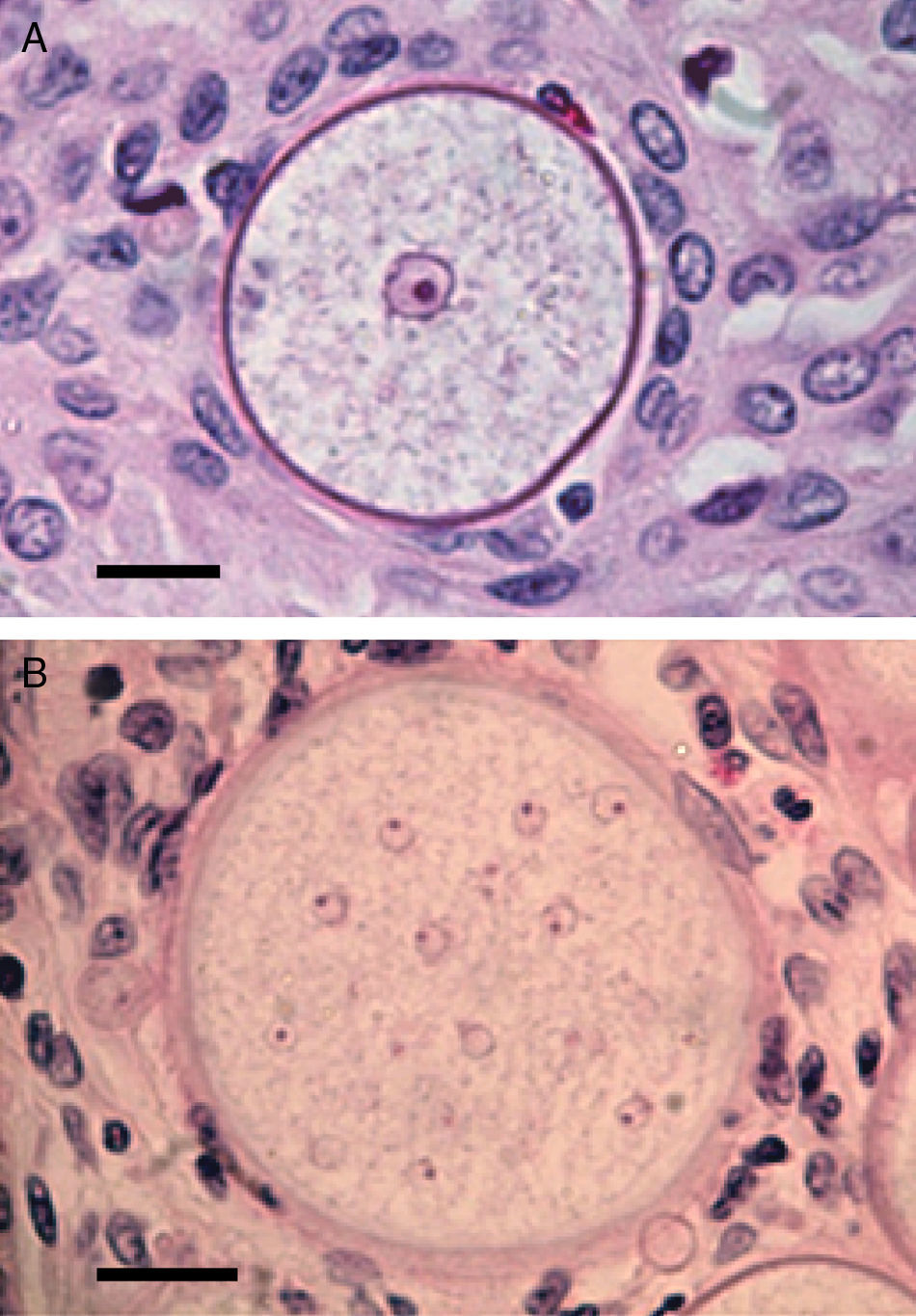
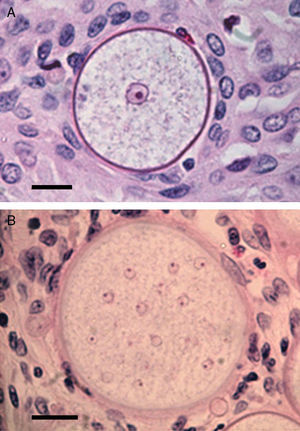
Panel A shows a histopathological section of Rhinosporidium seeberi stained with H&E depicting an early juvenile sporangium (JS) similar to the one depicted in Fig. 2 Panel B (Bar=8(μm). The presence of a well delimited cell wall, a granular cytoplasm with a typical reddish nucleus and a prominent nucleolus is typical of this stage. The JS are devoid of vesicles and EDBs, structures typically found in the endoconidia within or outside mature sporangia. The nucleus’ reddish color sharply contrasts with the hosts’ bluish nuclei surrounding the JS. Panel B depicts an intermediate sporangium with a thicker cell wall and several reddish nuclei, before nuclear division, identical to the one displayed by the JS in Panel A (Bar=20(μm). Note the contrast between R. seeberi's reddish nuclei and the host nuclei. This stage is characterized by multiple synchronized nuclear divisions.
Mature sporangia can reach 450μm or more in diameter (Fig. 1, this review). At this stage the sporangium is readily identified by the presence of hundreds of mature and immature endoconidia, by a thin cell wall containing at least three electron lucid and antigenic inner layers, the presence of an exit pore, and by its enormous size. The site of the exit pore (∼10–20μm in diameter) is believed to be genetically determined71 and is always facing the mature spherical endoconidia containing numerous vesicles, whereas the opposite site contains mostly small oval immature endoconidia (2–5μm in diameter) with few vesicles, and thus it has been referred by some as the “germinative zone” (Fig. 1, this review).87 Mendoza et al.,71 and others18,65,87 reported the formation of new endoconidia directly from this particular area. It has also been shown by Ashworth16 and Mendoza et al.71 that watery substances facilitate the release of mature endoconidia. These authors further proposed that upon contact with watery substances lytic enzymes are accumulated at the pore site, a feature that could facilitate the release of the endoconidia due to an increase in the osmotic pressure in the mature sporangia. This mechanism first proposed by Ashworth,16 was later adopted by Mendoza et al.,71 to explain the methodical release of the endoconidia from mature sporangia. A summary of R. seeberi life cycle, based on the accumulated thoughts and observations, is presented in Fig. 4 of this review.
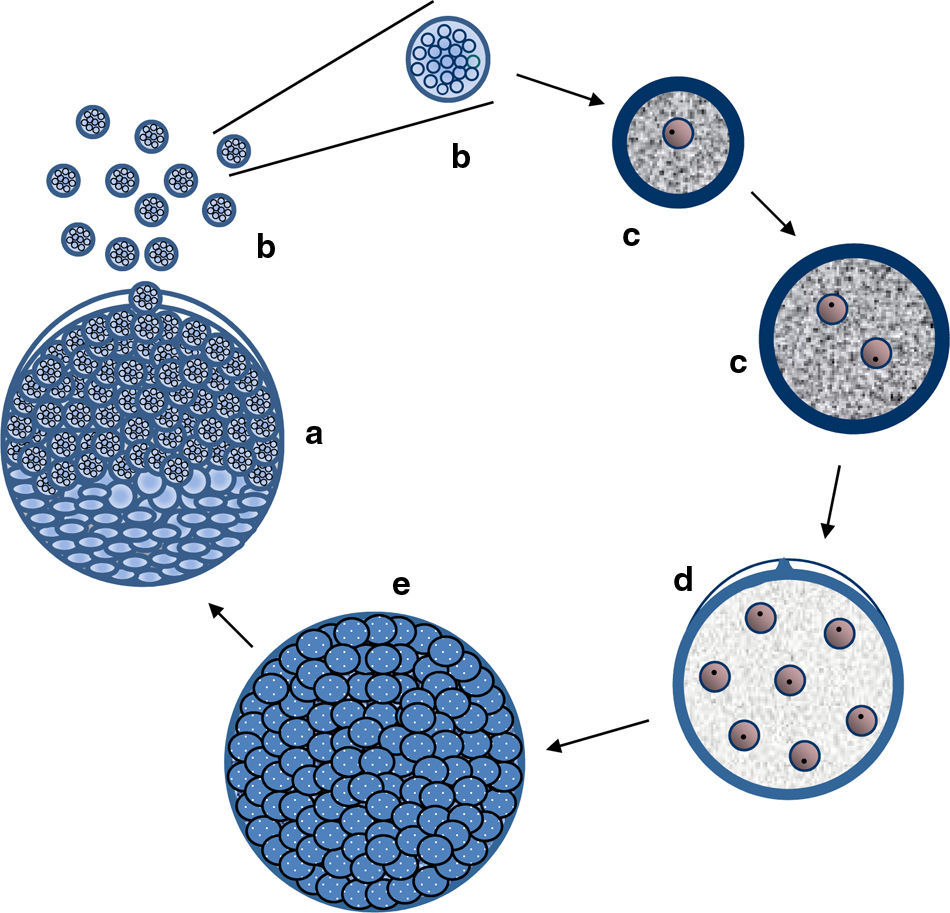
The figure depicts the parasitic life cycle of Rhinosporidium seeberi based on data published in the last 110 years. It starts with the release of the endoconidia through a pore from mature sporangia (a). The released endoconidia (b) increase in size (10–70(μm) loosing the morphological features typical of endoconidia such as vesicles and EDBs, becoming juvenile sporangia (JS) (c). This is the most commonly found phenotype in histopathological preparations. The JS are characterized by the presence of granular cytoplasm, reddish (in H&E) nuclei (could be a single nucleus or more than two nuclei) with a prominent nucleolus similar to those displayed in Fig. 3, and a thick cell wall. The JS increases in size (∼70–150(μm) and becomes intermediate sporangia (IS) (d). This is a rare encountered phenotype. The stage is characterized by synchronized nuclear divisions developing several hundreds of new nuclei without the formation of a cell wall within the sporangia. The sporangia at this stage possess a thick cell wall. The presence of rudimentary pore can be found at this stage, but it is a very rare event, thus difficult to find in histological sections. As the nuclei multiply, the IS continue increasing in size (≥150(μm) becoming an early mature sporangia (e). At this stage the last synchronized nuclear division takes place, after which a thin cell wall becomes apparent around each nucleus. Eventually, the sporangia reach maturity which is followed by the formation of a pore allowing the release of the endoconidia and the cell cycle is repeated (a).
Relevant to R. seeberi developmental features within the spherical cells are the long forgotten depictions of mitotically dividing nuclei and the presence of chromosomes, as reported by the early investigators.2,16,59 The presence of nuclei in the parasitic stages of R. seeberi was first recorded by Seeber,84 who on page 35 of his thesis depicted a nucleus within the “sporoblast” (endoconidium), a finding corroborated later by others.2,16,19,42,46,59,82,96,100 The first description of mitotic figures within immature sporangia, however, was that of Ashworth,16 who provided (Plate 1, Figs. 3–17) illustrations of the mitotic figures he observed of prophase and telophase nuclei during the first nuclear division and the presence of at least four chromosomes. The first comprehensive photographic description on the histological events of nuclear division within the JS and IS was presented in the thesis of Acevedo2 (pages 66–73), where he depicts several single nuclei and the first nuclear division within the endoconidia (Figs. 41, 42, 44). In addition, he also shows in Figs. 45–49 and 56–61 the presence of multiple nuclei and nuclear divisions within immature sporangia, similar to that depicted in Fig. 3B of this review. Acevedo's observations2 were validated six years later by Kurunaratne.59
In the first book ever published on “Rhinosporidiosis in Man” (an update of a previous edition), Karunaratne59 thoroughly reviewed all aspects related to R. seeberi including its parasitic cell cycle. Karunaratne59 showed photographs (Figs. 1–14) of the first nuclear division and the presence of four chromosomes (as predicted by Ashworth16) within the early JS and numerous nuclei within IS during prophase, metaphase and anaphase. With few exceptions, subsequent studies did not explore this fascinating feature of R. seeberi and mostly concentrated on other aspects of its life cycle.17,18,42,46,47,54,65,73,75,83,87,96,100,101
R. seeberi's eukaryote and prokaryote theoriesR. seeberi is a prokaryote cyanobacterium within the genus Microcystis. First Alhuwalia et al.,9 and then Alhuwalia6 reported that they have evidence indicating that R. seeberi is a cyanobacterium in the genus Microcystis. This hypothesis was unveiled in India based on ecological studies conducted in ponds and rivers where patients with rhinosporidiosis often bath. Environmental discoid and oval structures that, according to these authors, resembled the parasitic stage of R. seeberi in the infected tissues of patients with rhinosporidiosis were found. They stated that the cocci (nanocytes) developed by the genus Microcystis in these aquatic environments possessed striking morphological similarities with R. seeberi's endoconidia and thus, these structures may be the same elements observed in cases of rhinosporidiosis. They proposed that the nanocytes might be the infecting units of the diseases rhinosporidiosis. They also suggested that the amorphous and oval colonies filled with numerous internal cocci of Microcystis found in the ponds, were the same as mature sporangia with endoconidia observed in the infected tissues of patients with rhinosporidiosis. However, these observational data were not further investigated to validate the author's hypothesis.
The initial investigations putting forth the Microcystis hypothesis were followed by a study reporting its isolation using BG11 medium (to isolate cyanobacteria) and a slide culture technique with different concentrations of glycerol.9 According to the authors of this study described what they contend is a reliable method to culture R. seeberi. They also reported the isolation of Microcystis from clinical cases of rhinosporidiosis, and from water ponds, the detection of spherical structures containing nanocytes in BG11 medium and the formation of some filaments identified as non-fungal in origin in the slide cultures. Based on their findings, the authors concluded they had recovered from both, natural sources and clinical samples of patients with rhinosporidiosis, a cyanobacterium of the genus Microcystis and thus that this microbe was the legitimate etiologic agent of rhinosporidiosis.
Dhaulakhandi et al.,30 using molecular methodologies amplified a 16S rRNA sequence (accession number AJ440719) from purified single cells of R. seeberi obtained from patients with rhinosporidiosis and from environmental samples. The rRNA sequence contained the typical AATTTTCCG signature for cyanobateria and two other palindromic repeats present in Microcystis species, including Microcystis aeruginosa. Since the round bodies (RBs) of R. seeberi resemble Microcystis phenotypes and the 16S rRNA sequence has strong DNA identity with other cyanobacteria, they concluded that the etiologic agent of rhinosporidiosis is a cyanobacterium and thus, R. seeberi should be reclassified.70 More recently, Alhuwalia4 conducted new phylogenetic studies with the sequence obtained from their previous study and found that the 16S rRNA R. seeberi sequence has 98% identity with flowering plants and only 79–86% identity with Microcystis and with other cyanobacteria in the chroccocales. Despite this result, they argued that the morphological features displayed by R. seeberi in patients with the disease have more characteristics in common with the cyanobacteria than with the eukaryotes microbes in the Mesomycetozoa.4
In a letter to the editor, Alhuwalia5 defended her position arguing that the mistake leading Herr et al.42 to classify R. seeberi as a eukaryote microbe in the Mesomycetozoa, was the use of non-purified sporangia and endoconidia from the infected tissues of humans with rhinosporidiosis. She concluded that the amplified DNA sequence of Herr et al.,42 was probably of human origin, a claim properly refuted by the authors.70 She also stated that the structures in the EM figures identified by these authors as nuclei were “not nuclei but nanocytes of Microcystis encompassing naked prokaryotic DNA”. She further contended that the structures named as mitochondria by Herr et al.,42 were of human origin and not from R. seeberi sporangia. She based this conclusion on numerous EM studies that did not find mitochondria in young and mature sporangia.
R. seeberi is a eukaryote microbe within the Mesomycetozoa. In 1999 Herr et al.,42 provided taxonomic and molecular data indicating that R. seeberi is an eukaryote microbe within a new cluster of aquatic pathogens, first described by Ragan et al.84 as the DRIP group (after Dermocystidium, Rosetta Agent=currently S. destruens, Ichthyophonus, and Psorospermium), and renamed by Herr et al.42 as the Mesomycetozoa3 (Coanozoa of Cavalier-Smith23). This finding was welcome by the scientific community in part, because it corroborated 100 years of studies linking this pathogen to the eukaryotes.16–18,45–47,53,54,65,75,83,87,88,100,101 To reach their conclusion, Herr et al.,42 collected tissue samples from two Sri Lankan men with rhinosporidiosis and conducted EM and molecular studies. The EM studies of the infected tissues showed mature and immature sporangia of different sizes, some nuclei containing a prominent nucleolus and with numerous flat mitochondrial cristae detected in intermediate sporangia. Although this position was rejected as inaccurate by Alhuwalia5 (see above), in a response to her critique Mendoza et al.,70 provided additional EM photos substantiating their findings.
In the above study the tissue samples were processed in two ways: a) the sporangia were mechanically dissected from the infected tissues washed with sterile distilled water and then disrupted in the presence of glass beads, and b) the tissue samples were placed in a mortar and ground while frozen in the presence of liquid nitrogen with a pestle. The genomic DNA extracted from these samples was then subjected to PCR amplification using the universal primers NS1 and NS8.42R. seeberi 18S SSU rDNA 1790bp PCR DNA fragment was amplified (accession numbers: AF118851). Phylogenetic analysis of the 18S SSU rDNA sequence and 23 other DNA sequences of different microbes showed R. seeberi as the sister taxon to Dermocystidium species and closely related the members in the Ichthyophonida (see below).
Almost concomitantly, Fredricks et al.35 published an identical study claiming that they had sequenced the 18S SSU rDNA of R. seeberi from a dog with rhinosporidiosis. These investigators extracted genomic DNA from the same dog tissue sample used by Levy et al.65 In addition, they tested formalin fixed tissue samples of three humans with rhinosporidiosis, and nasal polyps from 12 human patients without the disease as negative controls. In this new study the design of several primers, including R. seeberi specific primers (Rhino-fw/Rhino-rev) based on DNA unique regions, were tested and a DNA biotinylated probe for fluorescent in situ hybridization (FISH) analysis was constructed. The results showed that the 18S SSU rDNA sequence amplified by Fredricks et al.,35 from the sample from a dog with rhinosporidiosis was identical to that reported by Herr et al.42 The specific primers designed to amplify only R. seeberi also failed to produce amplicons using genomic DNA of humans, fungi, Dermocystidium salmonis, and S. destruens (rosette agent). Moreover, the R. seeberi specific DNA FISH probe detected R. seeberi's DNA present in immature and immature sporangia (blue color), but the negative control probe showed only auto-fluorescence green (FITC) or red (Texas red) background according to the used wave-length filter. Of interest was the finding in EM of tubular mitochondrial cristae in R. seeberi's sporangia, a finding that contrasted with that of Herr et al.42 Nonetheless, the authors concluded that their study confirmed that R. seeberi is not a fungus, but instead it is an unusual aquatic protistan parasite located near the divergence between animals and fungi, thus validating the findings of Herr et al.42 They further stated that the specific primers and the FISH DNA probe they developed could be used to specifically detect this pathogen in putative cases of rhinosporidiosis.
R. seeberi is a fungus associated to the genera Synchytrium and Colletotrichum (Glomerella). In 2005 Thankamani99 announced that she has recovered in culture, from cases of rhinosporidiosis a fungal organism with “strong resemblance to the developmental stages of Synchytrium endobioticum, a member of chytridiales that causes black warts disease in potato”.99 In this preliminary report, Thankamani99 claimed that the same organism was consistently isolated from 15 swabs collected in 15 different Indian patients with rhinosporidiosis. The isolated organism (later labeled as UMH.48) grew very slow on blood and nutrient agar at room temperature, but failed to develop at 37°C. She described the colony recovered at room temperature as a very small (“pin tip size”) convex, circular, smooth, semitransparent colony without a diffusible pigment. When the primary isolate was subculture on Sabouraud dextrose agar (SDA) the formation of large sporangia with a “narrow pointing weak area, possibly, developing into a pore…” was noted.99 The author illustrates her findings with several figures showing the developmental stages of the spherical cells. These cells grew into spherical bridged cells containing mycelia-like structures and finally, by multiple divisions, developed into spherical spores. She stated that the formation of mycelia-like structures was a transient phenomenon leading to spore formation.99 After repeated subculturing, large sporangia could not be found. The formation of empty structures (prosorus-like, see below) was found attached to a homogeneous mass of nuclear material.
Thankamani99 reported also the presence of zoospore-like structures, but there were no description of their morphological features including the number of flagella and if the cells were motile. In Fig. 18 the author described “zoospores germinating/flagellated/sexual union of gametes forming zygospore”.99 However, since the flagellum cannot be Gram stained, the figure seems to display only some hyphal elements and a “spore” with a germ tube. She mentioned also that this strain was used to inoculate several mice, but none of the injected animals develop rhinosporidiosis. Later, Thankamani and Lipin-Dev98 investigated also the viability of R. seeberi and its developmental stages in a 10 year-old refrigerated sample. These authors claimed that they have isolated again the same organism (UMH.48) described earlier by Thankamani.99 They also illustrated with figures the findings encountered in this archival sample confirming Thankamani99 findings.
To validate the morphological data Thankamani and Lipin-Dev97 recently conducted molecular analysis using genomic DNA extracted from the original UMH.48 strain (kept on agar slants for 2 years) and directly from new biopsied tissues containing R. seeberi sporangia. However, the authors did not mention how many clinical samples were processed. They amplified the complete internal transcriber spacers (ITS) rDNA using the universal primers ITS5 and ITS4 in both the UMH.48 and the clinical samples.93 The authors were pleased to find out that the DNA sequence of the strain UMH.48 and that obtained from biopsied tissue sample with rhinosporidiosis shared 100% identity to each other. They went on to say that the “…99% identity between our isolate UMH.48 and the fungal DNA from rhinosporidiosis biopsy with respect to the 18S rDNA gene sequence categorically reaffirms that UMH.48 is the etiology of human rhinosporidiosis.”. Based on these data, they concluded that the strain UMH.48 seems to be a dimorphic fungus resembling Synchytrium species with the morphological features of R. seeberi as described by Seeber88 and Ashworth.16 When the sequences of both samples (UMH.48 and the biopsied tissue sample) were used to interrogate the database using BLAST analysis, 99% identity was found with the ascomycete Colletotrichum truncatum (Glomerella teleomorphic stage).97 The authors agreed that this finding was in direct contradiction with the morphological features reported on the original UMH.48 strain.99 But they did not elaborate more. Instead, they compared their sequences (accession numbers JN807465 and JN807466) with chytridiomycetes and mesomycetozoans sequences. As expected from the BLAST analysis, the sequences used by Thankamani and Lipin-Dev97 showed little identity in common with the members of these two groups of microbes. Although the authors did not conduct phylogenetic analysis with their DNA sequences, they stated that their study confirmed R. seeberi as a fungus.
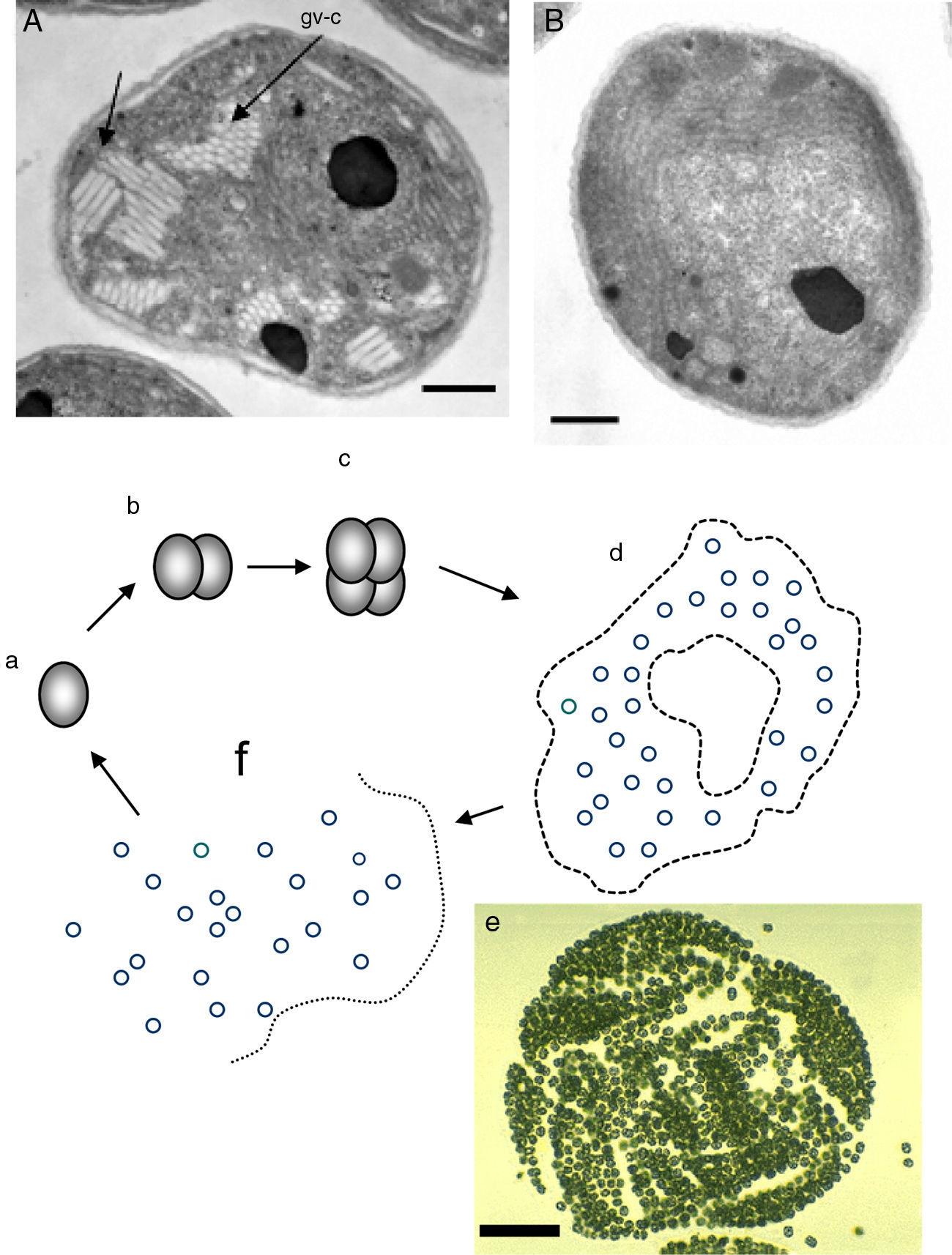
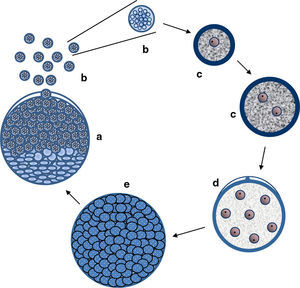
Critical review of R. seeberi's current interpretationsIs R. seeberi a cyanobacterium? The cyanobacteria comprise unicellular prokaryotic microbes currently grouped in five orders: Chroococcales, Pleurocapsales, Oscillatoriales, Nostocales, and Stigonematales.56,57 Members of the Chroococcales possess single spherical cocci (nanocytes) with homogeneous blue-green, grayish, or yellowish content and reproduce by binary fission in one, two, or three planes at right angles or in irregular planes (Fig. 5a–c, this review). Cells (cocci) can reach 0.5–30μm in diameter and form aggregate of nanocytes (colonies >200μm in diameter) displaying semispherical, discoid, or irregular shape formations depending on the planes of division (Fig. 5d and e, this review).80 The aggregated cocci (colonies) lack the presence of a true cell wall, but the colony secrets an extracellular slime that holds the nanocytes together. The reproduction of new colonies occurs by disintegration of the old colonies into small cluster of cells (Fig. 5f, this review) and new colonies are formed by the active aggregation of cocci (Fig. 5e, this review).57
Panel A shows EM photographs of Microcystis’ nanocytes with gas vesicles (gv) (arrows) (gv-l=long, gv-c=circular) a distinctive feature of the genus, whereas Panel B depicts a nanocyte without gas vesicles (Bars=500nm) (Courtesy of Dr. N. Tandeau de Marsac. Used with permission of J Bacteriol). The nanocytes with three peptidoglycan layers are observed in both panels. The lower section of Fig. 4 shows a cartoon demonstrating the typical binary fission division in more than one plane, typical of the genus Microcystis (a–c). The formation of large amorphous colonies by the aggregation of nearby cocci (d and e) is a common characteristic of the genus. The picture labeled “e” shows an actually colony of M. aeruginosa (Courtesy of Mark Schneegurt) (Bar=50(μm). Note an almost invisible mucilage matrix holding the nanocytes. Disintegration of old colonies (f) is necessary for the colonization of new environmental sites.
Members in the genus Microcystis, including M. aeruginosa, are classified within the Chroococcales.74,109M. aeruginosa is a common cyanobacterium found worldwide in fresh water environments. In nature M. aeruginosa cell division occurs by binary fission in three perpendicular planes and in a regular cubic arrangement of cocci (nanocytes), in the manner of those depicted in Fig. 5, this review.56,57,74,80 The nanocytes, by binary fission, develop into their final size (3–15μm) and shape (spherical) before the next cell division, but lack individual mucilage sheaths. Blue-green colonies are formed by the aggregation of several nanocytes forming spherical, oval, lobate or irregular colonies held together by a mucilage matrix (Fig. 5d and e, this review).57,79 Multiplication is by disintegration of the old colonies into cluster of nanocytes or single dispersed coccus.57,62,79,109
The cell developmental features of the genus Microcystis in nature are in contrast with the parasitic features of R. seeberi in infected hosts. For instance, in the infected host R. seeberi mature spherical cells, cleave their nuclei to develop numerous endoconidia (see above).37,39,45–47,73,100R. seeberi's endoconidia (5–10μm) are then released through a pore, and once in the infected tissues each endoconidium increases hundreds of times its original size (>450μm). The enlarged sporangia in turn produces hundreds of new endoconidia that are then released through a pore and the cycle is repeated (Fig. 4, this review).48,99 By contrast, in the genus Microcystis: a) nanocyte cell division occurs by binary fission in three perpendicular planes either outside or inside the colony's mucilage matrix; b) the shape and size of nanocytes remain unchanged throughout the entire cell cycle; c) in nature the amorphous or spherical colonies are formed by the aggregation of nearby nanocytes that became engulfed within a mucilage matrix; and d) the formation of new amorphous or spherical colonies takes place only after disintegration of old colonies. So far, there is not a single report describing: a) R. seeberi producing endoconidia by perpendicular binary fission in three planes; b) unchanging sizes of released R. seeberi endoconidia; c) formation of R. seeberi mature sporangia by the aggregation of nearby endoconidia; d) formation of an exogenous mucilage matrix in mature sporangia, or d) the disintegration of R. seeberi mature sporangia to form new mature spherical cells by aggregation. None of these Microcystis cell developmental features have been ever encountered in the tissues of the infected patients with rhinosporidiosis.2,16,46,47,54,59,63,75,87,101
According to Alhuwalia6 the morphological features of the genus Microcystis in nature and R. seeberi in the host's infected tissues are almost identical. However, a close inspection of the Microcystis nanocyte and the endoconidium of R. seeberi revealed contrasting differences. In addition to the presence of a distinctive nucleus in R. seeberi endoconidia as reported by many,2,16,40,54,59,100 Sleyter et al.,92 reported that Microcystis nanocytes in EM have three uniform peptidoglycan layers and an outer proteinaceous S-layer. In contrast, the endoconidia of R. seeberi lack these cell wall features (see above) and at this stage undergo dramatic changes to the thickness of its bilaminated cell wall, a feature absent in the genus Microcystis.62,92 Furthermore, the most distinctive feature of Microcystis’ nanocytes using EM is the presence of gas vesicles (GV) within the nanocytes (Fig. 5A, this review).44 This feature is considered almost diagnostic for the genus Microcystis, in which these structures play a key role during its life cycle in nature. In EM preparations GV are long conical or circular (depending on the angle of the section) hollow gas-filled structures44,74,101 (Fig. 5A and B, this review), which appear similar to empty bee-like honey combs in several places in a nanocyte. However, and in spite of having such unique identifying characteristics, no studies have documented the presence of entities similar to GV in R. seeberi endoconidia in patients with rhinosporidiosis.2,13,16,45–48,54,59,87,95–101 Although it could still be argued that the GV are only needed in nature and thus are not be displayed in other environments. Arguing against this particular possibility are the numerous other ultrastructural features of nanocytes devoid of GV (Fig. 5B, this review) that are equally different from those associated with R. seeberi endoconidia (Fig. 2A, this review).44,74,104 In summary, the reviewed literature indicates that R. seeberi's endoconidia do not divide into three planes and lack GV or GV-like structures.2,16,46,59,75,87,100,101 Although the intracellular vesicles in nanocytes, which are believed to be pools of reserve substances, have been also found in R. seeberi, these structures also possess key differences as well (Fig. 5A and B).45,47,62,101,104
The microscopic arrangement of the nanocytes in colonies of Microcystis is distinctively different when compared with the arrangement of endoconidia found in R. seeberi. Also, Microcystis colonies do not have a true cell wall and the nanocytes are held together by an amorphous mucilage matrix; whereas R. seeberi has a well developed thin cell wall, which contains three inner layers holding the endoconidia (Fig. 1, this review).2,16,42,54,59,71 In addition, R. seeberi mature sporangia with endoconidia are always characterized by their spherical shapes, whereas the aggregated colonies of the genus Microcystis display multiple morphologies depending on the environmental conditions.57,62,79,109 Together with the presence of a nucleus, these contrasting cell developmental differences distinctly differentiate Microcystis species from the genus Rhinosporidium.
Does R. seeberi possess rDNA from a cyanobacterium ancestor? The original proposal of Alhuwalia9 was based on the morphological attributes that Microcystis cells shared, according to this author, with R. seeberi. However, they did not perform EM studies to confirm their theory. Instead, a molecular study to prove that DNA of Microcystis sp. was present in the tissue of humans with rhinosporidiosis was carried out.31 This preliminary molecular study concluded that the DNA band pattern found in the water containing Microcystis cells matched the patterns observed from clinical samples and thus was used as proof of the “existence of a pathogenic strain of Microcystis for humans”.31 A drawback of this preliminary study was the lack of controls. Two years later Dhaulakhamdi et al.,30 published yet another molecular study. This time genomic DNA samples from: a) pure culture of M. aeruginosa, b) genomic DNA from putatively pure R. seeberi sporangia incubated in cell culture, and c) sporangia collected directly from clinical samples (at least 12 different clinical samples) were subjected to PCR-based analysis. They concluded that the amplified sequence has the signature for the 16S rDNA gene of cyanobacteria and thus, confirmed the prokaryotic nature of R. seeberi. However, when the amplified prokaryotic DNA from tissue samples infected with R. seeberi was compared to the homologous DNA sequence of M. aeruginosa only moderate identity was found. Sadly, the authors did not make further comments about this particular finding. Moreover, when the 16S rDNA deposited in the database was used in BLAST analysis the authors found 98% of identity with plastids present in flowering plants. This fact prompted Alhuwalia et al.,4 to justify that the relationship between R. seeberi 16S rDNA and those present in plants explain R. seeberi's typical auto fluorescence at 510–540nm wavelengths, a characteristic of photosynthetic microbes.
Based on ultra-structural studies depicting the presence of a true nucleus (see above) and the photographic evidence presented in this review, certainly R. seeberi is a eukaryotic microbe. If R. seeberi, in addition to being a eukaryotic microbe, possesses prokaryotic rDNA, then this finding is intriguing. Assuming that the original PCR experiments were free of environmental contaminants,30 this finding may be interpreted as the acquisition of prokaryote plastids by endosymbiosis; but does not necessarily means that R. seeberi is a cyanobacterium. On the contrary, this finding could provide new insights into the R. seeberi evolutionary history leading to the acquisition of such plastids from a cyanobacterium ancestor. Thus, endosymbiotic acquisition of a plastid in R. seeberi could be a possible explanation to this discrepancy. This scenario is plausible since the transfer of such plastids from ancient cyanobacteria ancestors to eukaryotic microbes have been well documented.28,33,36,56,86 So far, plastids transferred from cyanobacteria to eukaryotic cells have been found in algae,33 apicomplexans,51 cryptomonadas,29 dinoflagellates,36 and plants.28 If R. seeberi had acquired such plastids, it will be the first protist closely related to fungi and animals with this particular feature.
Is R. seeberi a eukaryote Mesomycetozoa linked to aquatic fish parasites? Although the presence of a nucleus is always a distinctive feature of eukaryotic organisms, the finding of this organelle to support a R. seeberi taxonomic link to the eukaryotes has proved to be difficult (see below) for various reasons: 1) because R. seeberi resists culture most studies have been conducted in the tissues of its infected host. Thus, depending on the tissue sectioning plane, the finding of a nucleus has been always fortuitous, 2) few investigators are familiar with the microscopic and ultra-structural characteristics of R. seeberi's nuclear division during its life cycle (see above), 3) the morphology of R. seeberi's nucleus and nucleolus in microscopic and EM preparations has yet to be properly defined, and 4) the presence of nuclei, mitochondria, nuclear membrane and other entities within the spherical structures of R. seeberi are directly affected by the protocols followed during formalin fixation treatment. Thus, investigators at different times have reached varied positions and applied different interpretations to their observations.2,9,16,47,53,54,59,64,75,87,101 However, with the advent of molecular methodologies a new door was opened. Good examples of the application of this new technology to the phylogeny of R. seeberi are the studies of Frederick et al.,35 Herr et al.,42 Pereira et al.,82 Silva et al.,91 and Suh et al.94 These independent teams all have reported that they have molecular data indicating that the studied epitopes of R. seeberi genomic DNA extracted from humans and animals, place this unique pathogen with previously unclassified protistan fish parasites. Perhaps the most important aspect of the finding was that two independent research groups, utilizing two different mammalian species with rhinosporidiosis, both amplified an 18S rDNA molecule sharing 100% identity, and both groups obtained identical phylogenetic trees.35,42 This finding leads both groups to believe that only one species, R. seeberi, was affecting the various animals having rhinosporidiosis.68
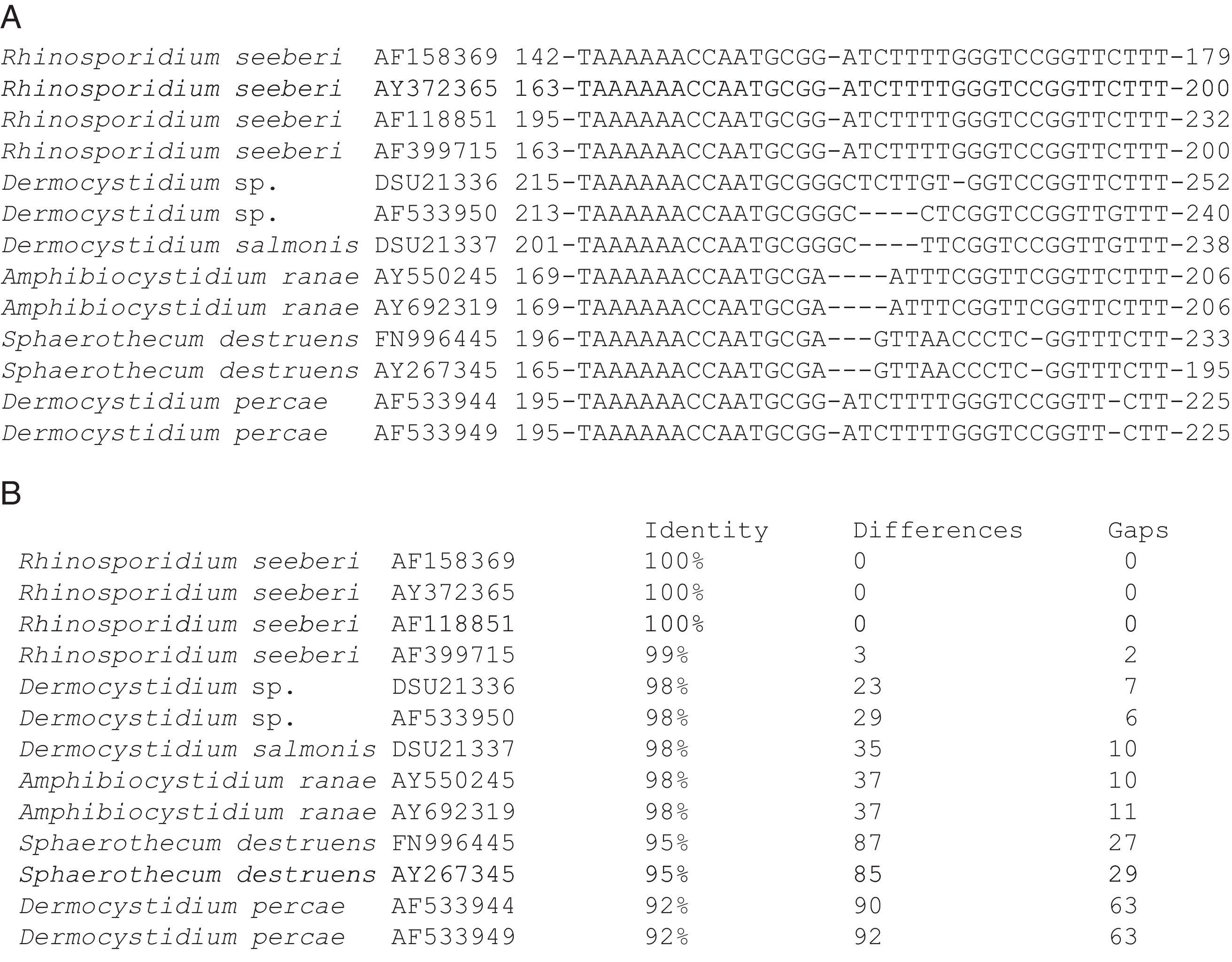
Alhuwalia4 stated that a strong argument against the eukaryotic theory of R. seeberi was that Herr et al.42 and Fredricks et al.,35 probably PCR-amplified DNA from endoconidia of the Dermocystidium spp. commonly found in aquatic environments.3,68 She predicted that the amplification of Dermocystidium 18S SSU rDNA was related to a few cells present in the tissues of humans and animals previously in contact with ponds containing putative fish infected with Dermocystidium spp. However, Fredricks et al.,35 clearly show that the specific Rhino-fw and Rhino-rev primers did not amplify D. salmonis genomic DNA used as a control, a datum that argues against this claim. Moreover, when the 18S SSU rDNA from D. salmonis spp. was used in BLAST analysis by us, key DNA differences were detected among these two groups of pathogens (Fig. 6, this review). Thus, the argument that the human tissue used as negative controls by Herr et al.,42 and Fredricks et al.,35 yielded negative PCR results, because the selected individuals had no previous exposure to environments containing spores of Dermocystidium, is untenable. Another finding that validates Fredricks et al.35 conclusions, is that their DNA in situ probe detected R. seeberi DNA inside the sporangia in histological preparations (blue color), but not in the negative controls (including D. salmonis), ruling out autofluorescence.90
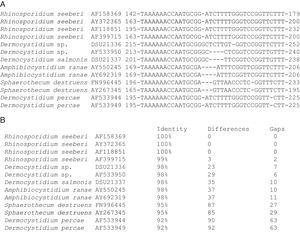
The partial 18S SSU rDNA sequences of several members in the Dermocystida are shown in Panel A. The differences that R. seeberi displayed at the DNA level when compared to Dermocystidium spp. and to the other members of the group are depicted. The DNA sequences accession numbers are placed after the species name. The numbers at the right and left section of each DNA sequences represent the location at molecular level of the selected epitope available at the NCBI database (Panel A). Panel B shows the percentage of identity, the numbers nucleotide differences, and the number of gaps encountered in the 18S SSU rDNA molecules of the 13 DNA sequences in panel A. The figure illustrates the differences encountered at DNA level between R. seeberi and the other members of the Dermocystida to support the concept7,8 that this mammalian pathogen can be differentiated using DNA sequence patterns from the genus Dermocystidum.
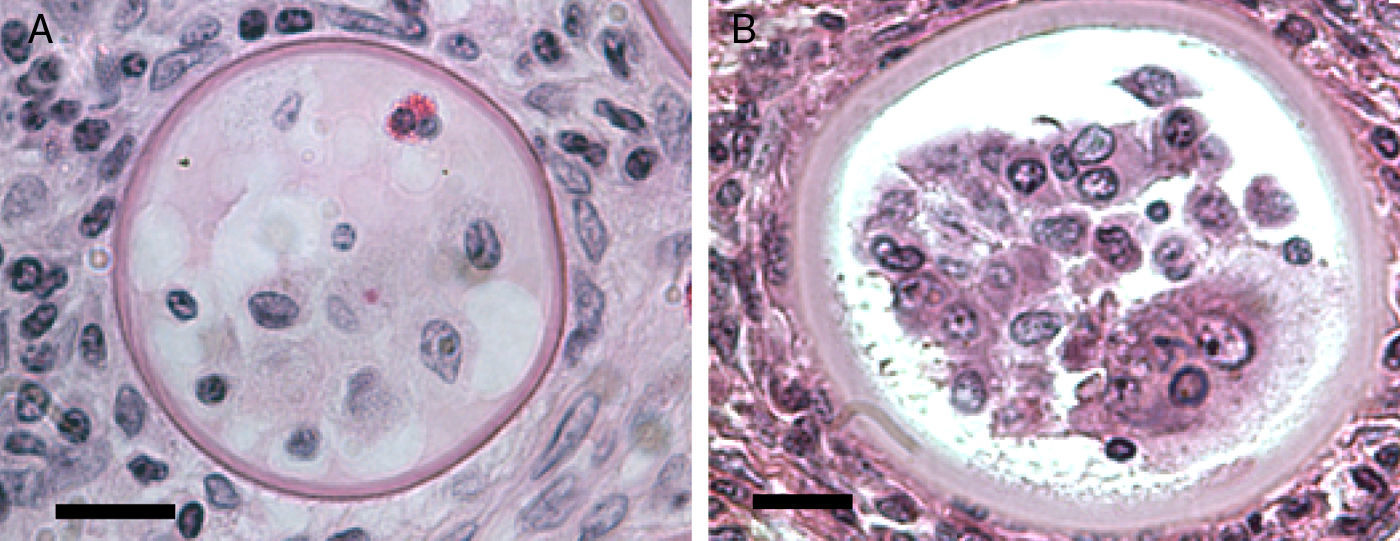
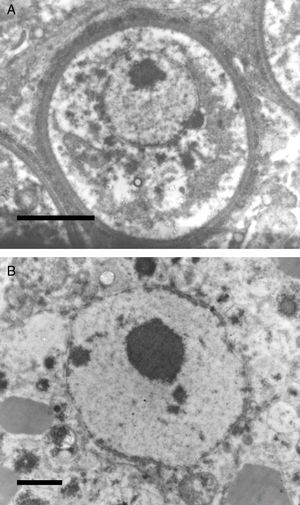
The EM Fig. 2 of Herr et al.,42 and Fig. 7 this review showed an IS with one or several nuclei and some mitochondria. Those supporting the prokaryotic nature of R. seeberi6,9 had argued that the nuclei and mitochondria depicted by Herr et al.,42 and that reported by others2,16,42,45,46,59,87 were more likely of human origin or “encompassing naked prokaryotic DNA”. These positions are understandable because of the common finding of inflammatory cells residing inside the cell wall of dead sporangia, such as the one depicted in Fig. 8 of this review. However, the description of the R. seeberi's nuclear features in Figs. 2 and 3 (this review) show substantial differences with the nuclei found in mammalian cells. For instance, Acevedo,2 Ashworth,16 and Kurunaratne59 described R. seeberi in H&E-stained preparations having a reddish spherical nucleus with a typical nucleolus surrounded by a delicate nuclear membrane similar to those depicted in Figs. 2 and 3 (this review). In multinucleated IS of R. seeberi (<150μm), nuclei with prominent nucleoli were identical to those found in single endoconidia and JS59 (Fig. 2A, this review). Alhuwalia5 arguments against the identity of the nucleus shown in the study of Herr et al.,42 were that these investigators confused a nucleus from the host tissue as R. seeberi's own nucleus. A close inspection of the host nuclei in histopathology (H&E) and EM (Figs. 2, 3 and 8, this review), however reveals that the host nuclei are strongly basophilic and their chromatin is condensed around the edges (Fig. 8, this review), whereas R. seeberi nuclei are smaller, pale, slightly eosinophilic and have a prominent nucleolus located at the center or slightly toward their edges (Figs. 2 and 3, this review).
The electromicrograph in panel A shows an enlargement of a juvenile sporangium with a typical nucleus and a prominent nucleolus (Bar=8.0(μm). Note the nuclear membrane surrounding R. seeberi's chromatin. Details of the nuclear membrane and the nucleolus can be observed in panel B (Bar=2.0(μm). In this EM close up the presence of several vesicles is also observed around the nucleus. Panels A and B and Figs. 2 and 3 (this review) give details on the typical morphological features of R. seeberi nuclei and establish without doubt that R. seeberi is a true eukaryote microbe.
Panels A (Bar=10(μm) and B (Bar=10(μm) show two different histological sections stained with H&E depicting dead sporangia of R. seeberi invaded by the host's inflammatory cells. The dead sporangia in panels A and B are commonly found in histological preparations. The nuclei observed inside the dead sporangia are indistinguishable from the nuclei of the host's inflammatory cells found around these sporangia (Panels A and B) and different to the R. seeberi's nuclei depicted in Fig. 3. The figure shows that R. seeberi nuclear morphological features can be properly differentiated from that of the host's own nuclei.
In Fig. 2B of this review the presence of three JS are depicted. This figure shows that the finding of a nucleus greatly depends on the plane of the sectioning. The JS on the left lacks a nucleus, whereas the other two sporangia show the presence of nuclei. The two JS on the right section of the figure depict the sectioning of the nucleus at different plains. Thus the nucleus present on the JS at the center appears smaller than it really is compared to the one that is closer to the right edge. More importantly, the presence of a macrophage nucleus at the top of the figure (arrow) clearly illustrates the contrasting differences between R. seeberi nuclei and in that of the infected hosts. Interestingly, the description of early investigators such as Acevedo,2 Kurunaratne,59 Kennedy et al.,54 and Thianprasit and Thangerngpol,100 all agree with the nuclear morphological features depicted by Herr et al.,42 in their Fig. 2, and thus supporting the interpretations of the nuclei described in this review (Figs. 2A, B, 3A, B, 7A, B, and 8A, B). We contend that Alhuwalia's5 argument about the presence of “encompassing naked prokaryotic DNA” in the pictures provided by Herr et al.,42 are unsubstantiated. Moreover, the chances that a circular prokaryote chromosome in an EM preparation could be sectioned in a way that shows the whole molecule in one section would be extremely difficult. In fact, it is more conceivable that the figures of Herr et al.,42 depicting several nuclei of different sizes surrounded by a nuclear membrane, comprise spherical structures sectioned at different points rather than a circular pattern entirely sectioned in one plane.
Is R. seeberi a pathogen that belong to the kingdom Fungi? The order Chytridiales (Chytridiomycetes) are divided into seven families: Olpidiaceae, Synchytriaceae, Achlygetonaceae, Rhizideaceae, Entophlyctaceae, Cladochytriaceae and Physodermataceae.49 Primitive species in this order are mostly holocarpic, whereas advanced species tend to be eucarpic, forming spherical cells.49 In the latter group, a system of rhizoids is present to anchor the cell to its host. The Chytridiales are intracellular pathogens with asexual reproduction by the development of uniflagellated planospores formed inside sporangia with one or two pore (papillae).49,51 Some species developed a typical cap at the tip of the pore termed operculum, but most species lack these opercula.49 In the latter case the flagellate spores are release through a pore in the cell wall or by the formation of an exit canal.49,52 In most species the sexual stage is yet to be found.49
The family Synchytriaceae, where Synchytrium species are located, comprises intracellular parasites forming uniflagellate planospores. They are holocarpic Chytridiales lacking opercula, a feature in common with the family Olpidiaceae.49–52 When a Synchytrium uniflagellated planospores reaches the infected hosts an amoeboid spore is formed and it sinks to the bottom of the parasitized cell where it germinates forming a structure termed prosorus.51,52 By nuclear divisions the prosorus is emptied and once outside, the cytoplasmic mass becomes the sorus with several multinucleated cells that at maturity transform into a sporangium with numerous planospores. It usually remains attached to the empty structure where the prosorus used to be located. If water is present, motile planospores are formed and released. The planospores have only one flagellum and exit the sporangium through a pore searching for a new host. Once they localize the host, the zoospores attach to the cell dissolving a section of the host's cell wall and penetrate leaving the flagellum outside the host.51 If water is scarce, the spores transform in planogametes and fuse in pairs to form zygotes.51,52 The genus Synchytrium has been divided by Karling52 in several subgenera, but they will not be discussed in this review.
The isolation of fungi from cases of rhinosporidiosis is not new. It has been recorded several times.14,58,59,69 The report of Thankamani99 is peculiar because the report mentioned the isolation of the same fungal organism from 15 different patients and from a ten-year old frozen sample.99 They described the isolation of a spherical fungus with all the morphological features that the author believes are very similar to that in the Chytridiales. The 24 figures describing the fungus in this report are not clear enough to confirm the author's claim. However, this is not the major problem of the report. When we compared the above discussed morphological features of R. seeberi with the description of Thankamani99 several inconsistencies were evident. First, there is not a single rhinosporidiosis report describing the formation prosorus-like structures emerging to form multinucleated sorus leaving behind empty structures (resting prosorus), to which it remains attached (as in the Chytridiales). Second, R. seeberi mature sporangia release endoconidia through a pore that do not develop flagella in the presence of water.16,71 In contrast, all chytridiomycetes in the presence of water release planospores through a pore with a single flagellum that is used to swim and find a new host. And third, Chytridiomycetes are mostly intracellular parasites, whereas R. seeberi cells cannot be intracellular in mammalian hosts by its enormous size. Moreover, the ultrastructural features of the cell wall in R. seeberi parasitic stages are different to that described in the members of the Chytridiales.53–57 And finally, the report of Acevedo,2 Ashworth16 and Kurunaratne59 of synchronized multiple nuclear divisions and the presence of mitotic figures in R. seeberi clinical samples are in direct contrast with the nuclear division and life cycle displayed by the Chrytridiomycetes and ascomycetes in their parasitic stages.53
Although Thankamani and Lipin-Dev97 stated that they have sequenced the 18S SSU rDNA of the UHM.48 strain, they amplified and worked with the ITS rDNA sequences. After BLAST analysis using the amplified DNA sequences of the original strain (UMH.48) and that recovered from a clinical sample, the authors were surprised to find out that their isolate was not a Chytridiomycete but an ascomycete in the genus Colletotrichum. This could well suggest that the morphological description of the original isolate was not accurate, or that the original strain contaminated later with Colletotrichum spores. Interestingly, the clinical sample amplified the same ITS DNA sequence as that in the UMH.48 strain. This was interpreted by the authors as the confirmation that this fungus is the etiologic agent of rhinosporidiosis. However, they did not discuss the possibility of cross-contamination between DNA samples, which could also explain the result.
In the discussion section the authors mentioned that those supporting the mesomycetozoa theory did not use the scientific method, but their own opinions on the subject to classify R. seeberi in the mesomycetozoa. However, the data generated by those teams are supported by multiple controlled phylogenetic analyses from numerous collected samples in humans and animals with rhinosporidiosis.35,42,87 The NCBI database contains many DNA sequences recovered from humans in India, Sri Lanka, the USA and Venezuela, as well as DNA sequences from several swans, dogs and cats with rhinosporidiosis. These sequences link R. seeberi with strong phylogenetic support to the mesomycetozoans. Therefore, the hypothesis that R. seeberi is a mesomycetozoa is based, no on a single case, but on scientific data collected by several independent teams from numerous mammalian and bird hosts with proven rhinosporidiosis.35,42,87,94
One of the main problems of those supporting the fungal theory of R. seeberi is the lack of controls. As per Mendoza et al.,69 the isolation of genomic DNA from uncultivated pathogens is difficult. Cross contamination with normal flora and environmental fungal and bacteria microbes is common.69 This had directed some investigators to misleading views about the taxonomy of well known uncultivated microbes.65 One of the controls missed by Thankamani and Lipin-Dev97 is the use of Fredricks et al.,35 specific primers for R. seeberi. This control would allow the authors to verify if the clinical sample under investigation possesses the same sequence as those reported in Indian human cases of rhinosporidiosis.91 This control would also suggest to the authors that the isolated fungal strain was or not a contaminant.
Two other topics mentioned by Thankamani99 deserve special attention. The UMH.48 strain was isolated on media commonly used in the laboratory: blood agar nutrient agar and SDA. The samples were incubated at room temperature, because the UMH.48 strain did not grow at 37°C. This strain also failed to infect mice. There are no reports describing a mammalian pathogen microbe that grows at room temperature but resists 37°C. Thus, based on this feature it could be concluded that the original strain was not a pathogenic fungus, but a common contaminant, probably Colletotrichum sp. Also, contrary to Thankamani and Lipin-Dev97 belief, Colletotrichum spp. are not dimorphic fungi as those studied in medical mycology. The authors supporting the fungal theory did not ask: why the early investigators failed to recover R. seeberi in culture? And why we succeed in 100% of the clinical samples? There seems to be a major inconsistency with the sudden success in recovering R. seeberi in culture. Thankamani and Lipin-Dev97 and Thankamani99 did not use new culture media or novel methodologies that have not been previously used by others. So, why other investigators had failed to culture R. seeberi using an identical approach? The answer seems to point more likely to an environmental contamination. So far all fungal isolates recovered from cases of rhinosporidiosis proved to be environmental fungal contaminants, and their apparent similarities shared with R. seeberi the lack of taxonomic expertise of those supporting these claims.14
The morphological and phylogenetic attributes of R. seeberi are in agreement with that displayed by the members of the Mesomycetozoa. The Class Mesomycetozoa, recently revised,68 comprises two Orders: Dermocystida and Icthyophonida. According to Herr et al.,42 and Fredricks et al.,35R. seeberi is located within the Dermocystida along with Amphibiocystidium, Amphibiothecum, Dermocystidium, and Sphaerothecum.3,68 The group is characterized by the presence of spherical unicellular microbes with the capacity to infect fishes, amphibians, birds and mammals.68 The main attributes of the group includes: 1) their typical spherical structures containing hundreds of endoconidia, 2) their long history of inclusion in different groups of microbes, including the fungi, 3) the resistance of some to culture, 4) all being aquatic microbes, and 5) all being pathogens of animals. But, the most striking similarity among them resides in their DNA make up. Phylogenetic analyses have shown that members of this group share similar evolutionary paths and together with members of the Ichthyophonida have a common ancestor located at the point where animals diverged from the fungal boundary.35,42,68,82
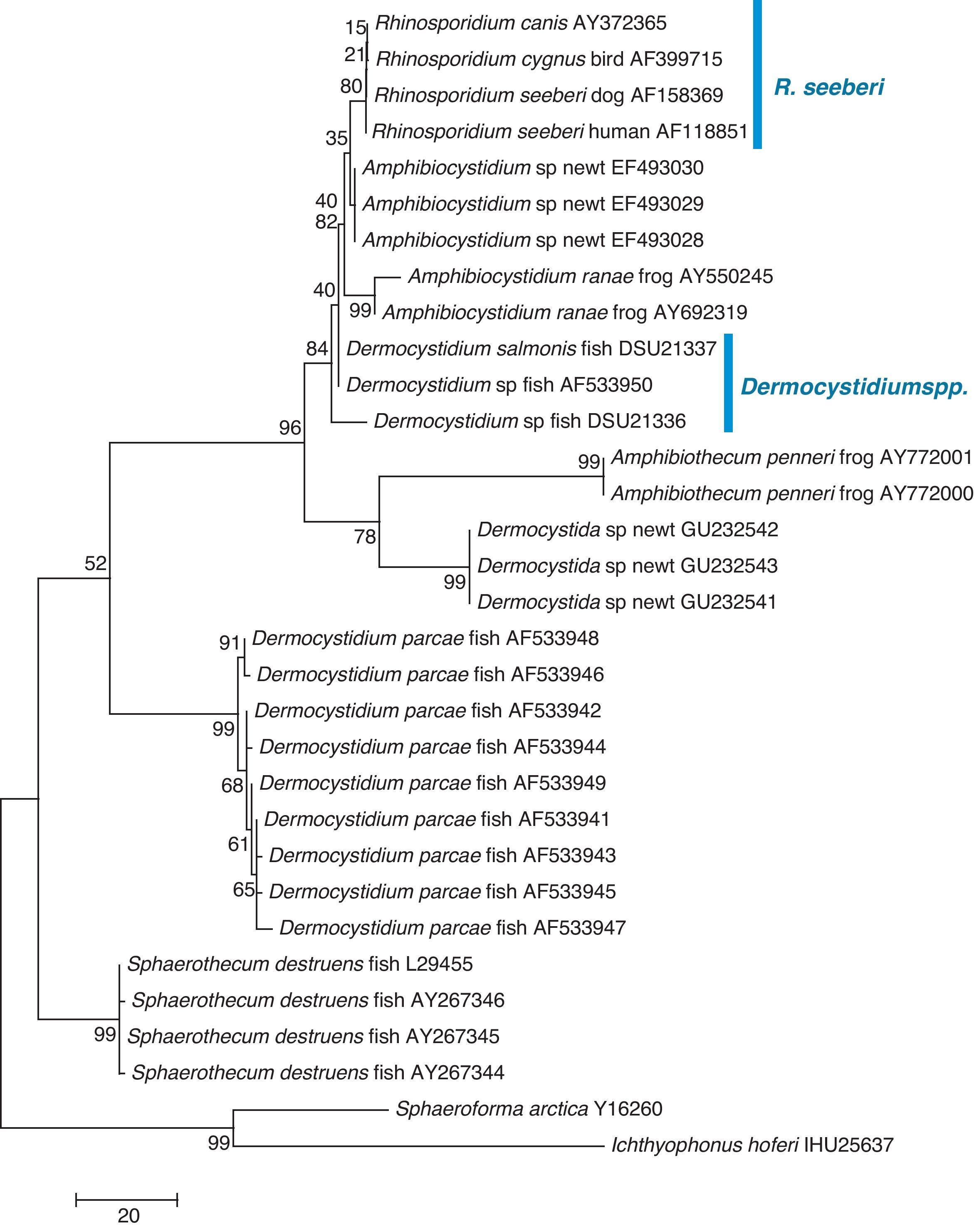
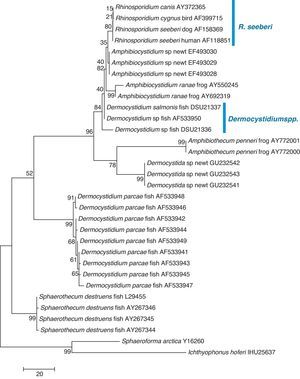
The finding that Ichthyophonus hoferi and R. seeberi both posses chitin synthase genes,41,93 as do other protistal microbes,76 was used to emend the term Choanozoa to Mesomycetozoa and also revealed another contrasting evolutionary feature that is not present in prokaryotic cells.3,56,68 In this context, phylogenetic analyses have shown (Fig. 9, this review) that R. seeberi forms a well supported sister taxon with A. ranae and with Dermocystidium species.82 This group in turn is sister to S. destruens and Dermocystidium percae. Pereira et al.,82 suggested that D. percae was probably not a Dermocystidium species, but a member of an entirely new genus, and proposed the name Dermothecum. Although the members in the Dermocystida shared similar DNA sequences, they nonetheless posses distinctive differences that are easily demonstrated using CLUSTAL and phylogenetic analysis as those shown in Figs. 6 and 9 of this review. This unequivocally demonstrates that the species of Dermocystidum and the other members of the Dermocystida can be properly separated using sequences alignment, phylogenetic analyses, and DNA probes. Taken together, these facts all further contradict Alhuwalia4,5 claims that Herr et al.,42 and Fredricks et al.,35 amplified DNA from samples putatively contaminated with Dermocystidium spp.
Parsimony tree aligned 18S SSU rDNA sequences of four R. seeberi and 28 other dermocystidian DNA sequences. The ichthyosporeans Sphaeroforma artica and Ichthyophonus hoferi were used as outgroup. Numbers above the branches are percentages of 1000 bootstrap-resampled data sets. The scale bar represents substitutions per nucleotide. The accession numbers are located after the organism's name. In this phylogenetic tree the four R. seeberi DNA sequences formed a well supported sister taxon to the five Amphibiocystidium spp. DNA sequences from newts and frogs. The three Dermocystidium species DNA sequences in turn formed a poor supported sister taxon to Amphibiocystidium and Rhinosporidium. The phylogenetic analysis indicates that Rhinosporidium seeberi shares more phylogenetic features in common with Amphibiocytidium than to the other members of the Dermocystida. Several strains of Dermocystidium percae were placed in a strongly supported taxon indicating that this pathogen of fish probably is not a Dermocystidium species. The genus Dermotheca has been suggested by some to differentiate this pathogen of fish from the other Dermocystidium species.
Mendoza et al.,68 and Pereira et al.,82 revised the microscopic and ultrastructural attributes of some members in the Dermocystida. The presence of nuclei very similar to those described by Acevedo,2 Kurunaratne,59 Savino and Margo,87 Thianprasit and Thangerngpol100 in humans, and Kennedy et al.,54 in swans infected by R. seeberi was also recorded by Pereira et al.,82 in frogs harboring A. ranae. Pereira et al.,82 showed a number of striking microscopic and molecular similarities that A. ranae shares with R. seeberi. Both pathogens not only develop spherules with numerous endoconidia, but both have identical ultrastructural features in common. For example, in Fig. 3A and B Pereira et al.,82 showed the morphological features of these pathogens in EM. They call attention to the fact that R. seeberi and A. ranae develop identical electron dense bodies (EDBs), and similar vesicles within the endoconidia. They also mention that the cell wall of the endoconidia and the mature sporangia of A. ranae share a number of features in common with R. seeberi. In panel B of Fig. 3 they show in both pathogens nuclei with a prominent nucleolus inside a mature endoconidium containing EDBs. They also stated that the presence of a nucleus in A. ranae endoconidia is very difficult to document, another interesting attribute in common with R. seeberi. In addition, S. destruens and Amphibiothecum penneri both develop spherical structures with endoconidia, and their ultrastructural features also share striking similarities with R. seeberi, but not with the Chytridiomycetes. Thus it is not surprising that Laveran and Petit63 and later Carini22 called the attention on the similarities of Dermosporidium from frogs (currently Amphibiocystidium) and R. seeberi.
In 2005 Silva et al.,91 studying the ITS DNA sequences of R. seeberi extracted from a dog in the USA, several humans in Sri Lanka and India, and at least two swans in Florida, reported that the phylogenetic analysis using these gene sequences suggested the possibility of several species specific strains within R. seeberi. The ITS DNA sequences of R. seeberi in this study again reinforce the concept that R. seeberi is indeed a eukaryotic pathogen with capabilities to infect mammals and birds. These phylogenetic analysis using the ITS sequences derived from the DNA of R. seeberi extracted from humans, swans, and dogs showed that they clustered in three strongly supported taxons, thus indicating the presence of at least three possible species-specific strains.
The discussed phylogenetic analyses and the revised microscopic and the EM morphological features of members in the Dermocystida are in agreement with that described in the parasitic life cycle of R. seeberi. Sadly, those involved with the prokaryotic and fungal theories did not conduct the appropriate microscopic and ultra-structural studies on their samples recovered from ponds, and R. seeberi from infected patients to avoid bias in the interpretation of their data.
R. seeberi is an eukaryote pathogen in the MesomycetozoaWhen Herr et al.,42 Fredricks et al.,35 and more recently Suh et al.94 found molecular data supporting the view that R. seeberi was an eukaryote pathogen sharing morphological and phylogenetic similarities with members of the Dermocystida, but distant from the fungi, their finding validated 100 years of similar views.2,16,18,24,45–51,54,59,75,85,87,88,100 More importantly members of the Mesomycetozoa are eukaryotic aquatic animal pathogens that manifest as spherical structures producing endoconidia, all features supporting the eukaryotic nature of R. seeberi.64 In contrast, there is not a single cyanobaterium or species of Chytridiomycetes that so far has been incriminated as causing disease in animals.21,43,56,57 The observations of a nucleus, as reported by many,2,16,18,24,38,45–47,54,59,75,85,87,88,94,100 and those depicted in Figs. 2, 3, and 7 of this review, are the most powerful argument against the hypothesis that M. aeruginosa is the etiologic agent of rhinosporidiosis. The revised microscopic and ultra-structural data2,16,18,24,45,53,54,59,75,85,87,88,100,108 and the figures showed in this review have repeatedly shown that Microcystis, Synchytrium and R. seeberi display fundamental morphological differences in histological preparations and EM studies. Thus, the claims that R. seeberi is microscopically similar to a cyanobacterium in the genera Microcystis and Synchytrium is scientifically untenable. In addition, we believe that the PCR amplification of cyanobateria DNA from tissue samples containing R. seeberi by Dhaulakhandi et al.,30 does not prove that this unique pathogen is a prokaryote. On the contrary, it may suggest that R. seeberi could have acquired plastids sometime during its evolutionary history. Delwiche et al.,28 using DNA sequences and phylogenetic analysis of the tufA gene have shown that the presence of plastids in the different eukaryote systems has all originated from cyanobacteria ancestors. Thus, the amplification of cyanobacteria DNA from clinical samples of patients with rhinosporidiosis,30 could imply that R. seeberi had acquired such plastids in the past.
Those involved in the fungal97–99 and the prokaryote theories4,6,8,9 make the same mistakes. Both groups isolated from clinical samples a microbe morphologically similar to R. seeberi, but failed to ask fundamental questions about the strength of their finding. The first question they should ask would be: what is the relationship of the investigated organism to similar microbes in the tree of life? Once that is determined, the most important issue is if the life cycle traits of these groups of microbes and their basic morphological features agree with that in R. seeberi. Both groups did not conduct ultrastructural studies on their organisms; instead they carried out DNA analysis directly from the isolated microbe and from clinical samples collected in patients with proven rhinosporidiosis. In both cases they claimed that the same DNA present in their isolate was also demonstrated in clinical samples and therefore, the isolated organism was the etiologic agent of rhinosporidiosis. This seems to be a new trend, so we have to be vigilant to avoid discrepancies and misinterpretation on future cases.
Finally, we considered of importance to mention that for the last 18 years those supporting the prokaryotic origin of R. seeberi have been also adopting multiple views on the nature of this anomalous pathogen. For instance, Alhuwalia in 19927 introduced the hypothesis that R. seeberi was “composed of both plant and human material that is self-assembled in response to specific function pertaining to its elimination from the tissue”. The same year Alhuwalia8 launched yet another theory. The author stated that “…The so-called sporangium is found to be a unique body containing residue-loaded lysosomal bodies (”spores“) for elimination of the system”. She also mentioned that “Two carbohydrates, namely defective proteoglycans synthesized intracellularly and an exogenous polysaccharide ingested through diet of tapioca constitute indigestible material in NB (nodular bodies) and scw (cellular waste)”. In 1994 Alhuwalia et al.,10 concluded that “…Dietary dry tapioca and chronic inflammation in undernourished individuals could lead to rhinosporidiosis”. Our careful review of these and similar concepts on the origin of R. seeberi by Alhuwalia and her colleagues suggest lack of the scientific rigor required to introduce novel concepts in science.
Concluding remarksWe have critically reviewed key microscopic, ultrastructural, and the molecular studies of the past 110 years and concluded that R. seeberi: i) does not share the microscopic or ultrastructural features with that found in the genera Microcystis or Synchytrium spp., ii) has entirely different life cycle traits than those reported for the genus Microcystis the chytridiomycetes or the ascomycetous fungi, iii) possesses a nucleus with a prominent nucleolus and distinctive features distinguishable from those associated with the nuclei of its infected hosts, iv) is a mesomycetozoa microbe based on DNA and phylogenetic analysis, v) has microscopic and ultrastructural characteristics that are in agreement with the characteristic of the Mesomycetozoa, but not with that of the members of lower fungi or ascomycetes vi) develops mitotic figures in prophase, metaphase, and anaphase without cytokinesis in its immature sporangia and, thus it is a typical eukaryote microbe taxonomically and phylogenetically different from the fungi, vii) may have acquired plastid from a cyanobacterium ancestor, and viii) does not develop uniflagellate cells in any of its developmental stages.
One of the fundamental features discussed at length in this review is the presence in R. seeberi of a typical nucleus and the reports of mitotic figures in tissue sections by the early investigators.2,16,59 The finding of synchronized nuclear division with the formation of endoconidia only in the latest mature stages, supports the placement of this unique pathogen in the mesomycetozoa and away from the fungi.2,16,59 We anticipate that the notion of plastid DNA in R. seeberi will ignite a new interest in the subject, which could redirect future research efforts in this and other areas that still need further research.
Conflict of interestThe authors declare no conflict of interest.
The authors thank Dr. Paul Szaniszlo for the suggestions and constructive criticism. The study was supported in part by the Biomedical Laboratory Diagnostics, Michigan State University.