The extraction techniques of deoxyribonucleic acid (DNA) from old bone samples are based on the use of complex methodologies, which deal with the degradation of the sample, the low amount of DNA, and the presence of inhibitors that can be extracted simultaneously.
ObjectiveTo compare the efficiency in obtaining DNA from the methods: organic extraction, commercial kit, and previous total demineralization, to obtain STR profiles of old bone samples from 3 taphonomic processes (alkaline, acid, and moist mitotic).
Materials and methods29 skeletal remains from 3 different taphonomic contexts were processed: acidic, alkaline, and wet-fungal. The amount of DNA obtained from 3 methodologies was evaluated in a comparative manner: organic extraction method (phenol–chloroform–isoamyl alcohol) (SO), extraction by silica column KIT QIAamp® DNA Investigator of QIAgen® (KC), and the methodology of extraction by previous total demineralization (DP). Finally, the obtaining of the STR profiles from the methodology of greater performance was tested.
ResultsThe following quantification values were obtained: (i) alkaline medium: 0.068±0.07 ng/μL (SO), 0.021±0.01 ng/μL (KC), and 0.073±0.052 ng/μL (DP); (ii) acidic medium: 0.098±0.064 ng/μL (SO), 0.041±0.029 ng/μL (KC), and 0.068±0.042 ng/μL; (iii) wet-mitotic medium: 0.25±0.061 ng/μL (SO), 0.04±0.027 ng/μL (KC), and 0.15±0.072 ng/μL (DP). Likewise, using the DNA samples obtained by the SO method, complete profiles were obtained for the wet taphonomic context while the alkaline taphonomic process proved to be the most drastic for DNA degradation, presenting a greater number of incomplete profiles.
ConclusionsThe methodology of organic extraction was optimal in obtaining DNA from the 3 taphonomic processes evaluated. On the other hand, the wet-fungal taphonomic process is the one that produces the least negative impact on the preservation of DNA from skeletal remains.
Las técnicas de extracción del ácido desoxiribonucleico (ADN) a partir de muestras óseas antiguas se basan en el uso de metodologías complejas, que lidian con la degradación de la muestra, la escasa cantidad de ADN y la presencia de inhibidores que pueden extraerse simultáneamente.
ObjetivoComparar la eficiencia en la obtención del ADN de los métodos: de extracción orgánica, kit comercial y de desmineralización total previa, para la obtención de perfiles STR de muestras óseas antiguas procedentes de tres procesos tafonómicos (alcalino, ácido y húmedo mitótico).
Materiales y métodosSe procesaron 29 restos óseos procedentes de tres contextos tafonómicos diferentes: ácido, alcalino y húmedo - micótico. Se evaluó, de manera comparativa, la cantidad de ADN obtenida a partir de tres metodologías: método de extracción orgánica (fenol-cloroformo-alcohol isoamílico) (SO), extracción mediante columna de sílica Kit QIAamp® ADN Investigator de QIAgen® (KC) y la metodología de extracción por desmineralización total previa (DP). Finalmente se ensayó la obtención de los perfiles STR a partir de la metodología de mayor rendimiento.
ResultadosSe obtuvieron los siguientes valores de cuantificación: i) medio alcalino: 0.068±0.07 ng/μL (SO), 0.021±0.01 ng/μL (KC) y 0.073±0.052 ng/μL (DP); ii) medio ácido: 0.098±0.064 ng/μL (SO), 0.041±0.029 ng/μL (KC) y 0.068±0.042 ng/μL; iii) Medio húmedo-mitótico: 0.25±0.061 ng/μL (SO), 0.04±0.027 ng/μL (KC) y 0.15±0.072 ng/μL (DP). Asimismo, empleando las muestras de ADN obtenidas por el método de SO, se obtuvieron perfiles completos para el contexto tafonómico húmedo en tanto que el proceso tafonómico alcalino evidenció ser el más drástico para la degradación del ADN, presentando mayor cantidad de perfiles incompletos.
ConclusionesLa metodología de extracción orgánica resultó óptima en la obtención del ADN a partir de los tres procesos tafonómicos evaluados. Por otro lado, el proceso tafonómico húmedo-micótico es el que menos impacto negativo produce en la preservación del ADN a partir de restos óseos.
In forensic genetic analysis, expertise in obtaining genetic material is a determining factor in solving enforced disappearances following acts of war and crime, and in identifying victims of natural disasters. Information obtained from DNA provides a more objective investigation, enabling the identification of the victims and perpetrators, constituting a basis for the criminal and social justice to which victims and their families aspire.1 However, although there are a variety of DNA sources, few are able to preserve the stability of this macromolecule over time.
As soon as the cell dies, a series of biochemical processes are initiated that lead to degradation of the cell components including the nucleus, where the genetic material is stored, ending with the degradation of the biological sample.2 These processes are very rapid in soft tissues; however, they slow down in hard tissues such as bone, due to its rigid structure and specific components that stabilise DNA (such as hydroxyapatite).3,4 Therefore, the success of genetic results, according to the location of the bone elements, is the cornerstone of any forensic investigation of war and criminal acts, such as enforced disappearances, wars, ethnic conflicts, terrorist activities, organised violence, among others. This investigation is generally conducted many years after the conflict, and therefore the information is less and more hidden (completely skeletonised, highly degraded, and conglomerates of skeletal remains).
Obtaining DNA from ancient human skeletal material depends on its own structure and on the particular conditions of burial or the environment in which the material was deposited, which influences the degradation process of particular remains.1 The conditions that most influence DNA degradation (taphonomic conditions) include temperature, humidity, oxygen levels, microorganism activity, pH, and/or the presence of certain compounds in the soil that are powerful inhibitors.1 The main purpose of forensic taphonomy is to detect environmental impact that has affected the skeletal remains, and thus design the best techniques to recover genetic material.5
The ideal technique for quantifying DNA from forensic samples is real-time PCR (RT-PCR), a procedure that characterises the DNA quantitatively and qualitatively, revealing the total amount of DNA present in the extracted samples, the presence of inhibitors, and male DNA. This information will help establish the optimal amount of DNA (depending on the state of each sample) to be used in the amplification stage. Because very small amounts of DNA can be amplified (from 0.5 to 2 ng/μL by PCR), the sensitivity of this technique has been established,6 and it is therefore recommended for the analysis of skeletal remains whose DNA is found in very small amounts, making it possible to continue with forensic investigations, where other techniques can also be used in the final identification.7
The aim of the present research study is to contribute with knowledge, based on 3 extraction methodologies used in the handling of skeletal remains derived from different taphonomic processes, to obtain a genetic profile. It should also be noted that our study does not seek to identify the skeletal remains, but rather to demonstrate the capacity of these 3 methodologies for their subsequent use in forensic practice in Peru.
Materials and methodsSamplesTwenty-nine bone samples were processed for this descriptive and retrospective study. These samples, which were associated with cases against humanity, were in the custody of the Public Prosecutor's Office in Ayacucho and were sent to the Molecular Biology and Genetics Unit of the Institute of Legal Medicine of the Public Prosecutor's Office in Lima, Peru (with ISO 9001 certification level) in 2009 and 2012. The sampling dates were from 1983 to 1985, which establishes an approximate age range of the samples of 25–29 years. The remains were kept at 4 °C until processing in 2014.
Taphonomic processes identifiedAlkaline medium: 15 exhumed samples belonging to the case “Cabitos (CAB), La Hoyada” (Department of Ayacucho). The soil pH is 7.9–8.5, sandy, with sparse vegetation, ambient temperature of 20–25 °C. Macroscopically, the skeletal remains show a surface with little corrosion, with noticeable changes in colouring, bone loss, cracking, and flaking.
Acid medium: 8 burial samples from the “Pichari” case (PIC) (Department of Cusco). Acidic pH environment 5.5–6.0, ambient temperature 6–8 °C, and dense vegetation. Bone remains with heterogeneous surface from smooth and homogeneous to areas with corrosion, cracking, flaking, and root marks.
Wet-fungal environment: 6 burial samples from the “Pucayacu” case (PUC) (Department of Ayacucho). Sandy soil pH 7.0–7.7; relative humidity 89%, fluvial climate, low and constant temperatures, and low-density vegetation. Macroscopically, the bone remains show corrosion, slight flaking, and the bodies also show evidence of adipocere with fungal growth.
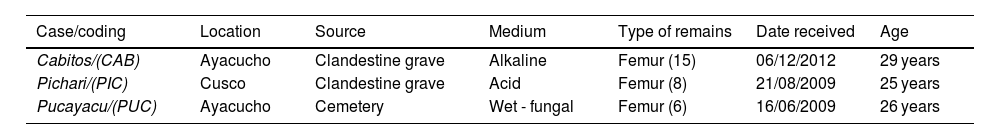
The types of samples for each medium are specified in Table 1.
Samples used in the present study.a
| Case/coding | Location | Source | Medium | Type of remains | Date received | Age |
|---|---|---|---|---|---|---|
| Cabitos/(CAB) | Ayacucho | Clandestine grave | Alkaline | Femur (15) | 06/12/2012 | 29 years |
| Pichari/(PIC) | Cusco | Clandestine grave | Acid | Femur (8) | 21/08/2009 | 25 years |
| Pucayacu/(PUC) | Ayacucho | Cemetery | Wet - fungal | Femur (6) | 16/06/2009 | 26 years |
First, the surface layer of the samples was scraped to remove exogenous DNA. The samples were then exposed to UV light for 15 min per side. For pulverisation, we followed the procedure described by Adler et al.8 using a Dremel electric drill at a slower speed (1000 rpm). Once the bone powder was obtained, the initial amount of 1.5 g was weighed and coded to proceed to the next step.
DNA extractionOrganic extraction method (Phenol:Chloroform:Isoamyl Alcohol): We followed the methodology of Miller et al.9 modified by Ausubel et al.10 Briefly, 1–1.5 g of the bone powder was placed in a 50 ml Falcon tube with 6 ml of digestion buffer (10 mm Tris–HCl, 0.1% SDS, 50 mm EDTA, 39 mm DTT) and 400 μL of proteinase K (20 mg/ml) and incubated in a water bath at 56 °C overnight. This was then centrifuged at 4000 rpm for 5 min and the supernatant was transferred to another 50 ml Falcon tube to be mixed with a phenol:chloroform:isoamyl alcohol solution in a 25:24:1 ratio, respectively. The mixture was centrifuged at 14 000 rpm for 3 min and checked for clarity of the aqueous phase, if not, the mixture was run again for clarification. A total of 400 μL of the clean aqueous phase, containing the DNA, was transferred to a Centricon® YM-100 column (previously hydrated with 400 μL of nanopure water) and centrifuged at a speed of 3500 rpm for 10 min, and the rest of the aqueous phase was centrifuged in the same way. Finally, the retained DNA was eluted by inverting the column and adding a volume of 400 μL of nanopure water and centrifuged at a speed of 3500 rpm for 10 min. The resulting DNA was stored at 4 °C for immediate use or at −20 °C for later use.
Extraction methodology by silica column extraction QIAamp® DNA Investigator Kit from QIAgen®: We followed the manufacturer's instructions. Briefly, 100 mg of the pulverised sample was placed in a 1.5 ml tube to which 360 μL of ATL buffer and 20 μL of proteinase K were added, the mixture was incubated overnight at 56 °C. Then, 300 μL of AL buffer was added by vortexing for 10 s, incubated at 70 °C for 10 min and centrifuged at 8000 rpm for 10 min. The supernatant was transferred to a 1.5 ml vial and mixed with 150 μL of absolute ethanol. The mixture was transferred to the QIAamp MinElute column to centrifuge at 8000 rpm for 1 min, and washed successively with 600 μL of buffer AW1, 700 μL of buffer AW2, and 700 μL of absolute ethanol centrifuging in each case at 8000 rpm for 1 min. The lid was left open at 56 °C for 3 min. Finally, it was centrifuged at 14 000 rpm to remove any residue and coupled to a new 1.5 ml tube. DNA was eluted by adding 50 μL of molecular grade water and centrifuging at 14 000 rpm for 1 min. The filtrate was stored at 4 °C until use.
Extraction methodology by previous total demineralisation and QIAamp® DNA Investigator silica column from QIAgen®: We followed the procedure described by to Loreille et al.11 Briefly, in a 50 ml Falcon tube, 1 g of pulverised bone, 15 ml of 0.5 M EDTA, and 1.5 ml of 10% SDS were placed in proportion in a 50 ml Falcon tube. The mixture was homogenised and incubated in a 56 °C water bath overnight. The mixture was centrifuged at 4000 rpm for 10 min and the sediment was incubated with 6 ml of lysis buffer (10 mm Tris–HCl, 0.5 M EDTA, 0.1% SDS, 39 mm DTT) in a water bath at 56 °C for 24 h. The mixture was then centrifuged at 4000 rpm for 10 min. It was then centrifuged at 4000 rpm×10 min and the supernatant was concentrated using Amicon Ultra centrifugal filter units Ultra-4, 30 kDa MWCO tubes, to a volume of 2 ml, with which the DNA was purified using QIAamp® MinElute silica columns from the QIAamp® DNA Investigator commercial kit from QIAgen® according to the above methodology. Once the DNA was recovered, the samples were stored at 4 °C until use.
DNA quantification by real-time polymerase chain reaction (RT-PCR)DNA quantification was performed with the Quantifiler Human model and the Quantifiler Y-chromosome model, using the Applied Biosystems® Quantifiler® Duo DNA Quantification kit, following the manufacturer's recommendations, using the Applied Biosystems® ABI PRISM SDS 7500 Real-Time PCR Systems Genetic Analyser; using a standardised in-house real-time PCR protocol. Both models rely on the amplification of a specific region of the human ribonuclease gene (14q11.2, 140 bp) and the SRY gene (Yp11.3, 130 bp) respectively, in addition to an internal PCR control (IPC). The master mix was prepared on the QIAgility automated robot from QIAgen®. The following amplification programme was used: 50 °C for 2 min; 95 °C for 10 min; 40 cycles of 95 °C for 15 s, and 60 °C for 1 min.
In addition, a positive control (pre-standardised quality DNA) and a negative control were considered, the latter containing the PCR master mix, but without any DNA template (which is replaced by ultrapure water), which in turn is used as external contamination control.
Analysis of the presence of inhibitors and clean-upBased on the IPC value, which is provided by the Applied Biosystems Quantifiler® Duo DNA Quantification kit, if a quantification value has not been obtained (indeterminate value), 2 options could be considered: (1) that a sample has no DNA, if the IPC value <30; or (2) that the sample presents PCR inhibitors, if the IPC value >30; for the latter case, the DNA was further purified using QIAamp® MinElute silica columns from the QIAamp® DNA Investigator commercial kit from QIAgen® according to the manufacturer's instructions.
DNA quality analysisThe quality of the products was assessed by obtaining genetic profiles based on autosomal STR (Short Tandem Repeat) markers. The commercial AmpFℓSTR® IdentifilerTM PCR Amplification Kit from Applied Biosystems was used for 16 STR markers (15 autosomal STR microsatellite markers plus 01 marker for sex determination), in a Mastercycler® Gradient-Eppendorf thermal cycler. Capillary electrophoresis and fluorescence detection using the ABI PRISM® 3500 Automatic Genetic Analyzer from Applied Biosystems was used to detect amplified products. The observation of peak size (above 50 RFU) was contrasted with standard reference markers, assuming that the negative control and the blank did not present any peaks.
Statistical analyses were performed on a personal computer using SPSS version 11.5 for Windows. Values are expressed as mean values±standard deviation. P-values less than .001 (P<.001) were considered statistically significant.
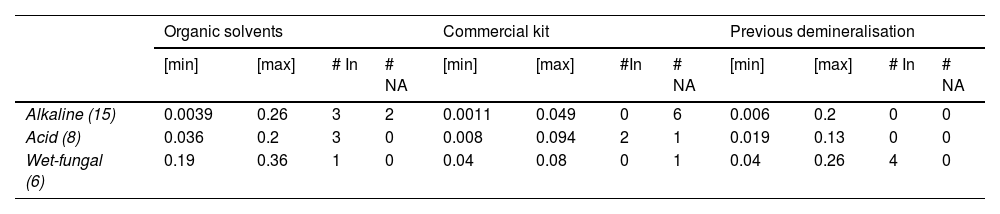
ResultsTable 2 shows the amount of DNA retrieved by RT-PCR. This procedure was performed on 29 bone samples, the results of which show that DNA was obtained from most of them.
Summary of DNA retrieval from different taphonomic processes using the 3 extraction methods.
| Organic solvents | Commercial kit | Previous demineralisation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [min] | [max] | # In | # NA | [min] | [max] | #In | # NA | [min] | [max] | # In | # NA | |
| Alkaline (15) | 0.0039 | 0.26 | 3 | 2 | 0.0011 | 0.049 | 0 | 6 | 0.006 | 0.2 | 0 | 0 |
| Acid (8) | 0.036 | 0.2 | 3 | 0 | 0.008 | 0.094 | 2 | 1 | 0.019 | 0.13 | 0 | 0 |
| Wet-fungal (6) | 0.19 | 0.36 | 1 | 0 | 0.04 | 0.08 | 0 | 1 | 0.04 | 0.26 | 4 | 0 |
# In: Number of samples that showed inhibitors (IPC Ct= > 31); max: maximum concentration obtained (ng/μL); min: minimum concentration obtained (ng/μL); # N.A: Number of samples where no amplification was obtained.
Fig. 1 shows the quantification levels obtained in each taphonomic process using the 3 extraction methods. The wet-fungal taphonomic process had the least negative impact on the recovery of genetic material, showing the highest minimum and maximum DNA concentrations compared to the other 2 processes (significance level <.001, Student's t-test). Also, in this taphonomic process, only 1 sample, which was processed with the commercial kit methodology, was negative for DNA retrieval.
The average quantification values for each taphonomic process obtained were: (i) for the alkaline medium: 0.068±0.07 ng/μL (OS), 0.021±.01 ng/μL (CK), and 0.073±0.052 ng/μL (PD); (ii) acid medium: 0.098±0.064 ng/μL (OS), 0.041±0.029 ng/μL (CK), and 0.068±0.042 ng/μL; (iii) wet-fungal medium: 0.25±0.061 ng/μL (OS), 0.04±0.027 ng/μL (CK), and 0.15±0.072 ng/μL (PD). However, outliers were shown in the alkaline medium using the SO (0.26 ng/μL) and PD (0.2 ng/μL) methods (Fig. 1). The heterogeneity of the values obtained is evident in the standard deviation data analysed and is due to the amplitude of the minimum and maximum values obtained (Table 1).
With respect to the presence of inhibitors, there is no direct relationship between the number of samples affected and the taphonomic process. In the acid and wet-fungal taphonomic processes, 5 samples showed the presence of inhibitors (IPC Ct=>31); the acid process was where the presence of inhibitors was detected for the 3 methodologies. However, when correlating the presence of inhibitors with respect to the extraction methodology, in the method using organic solvents, samples with the presence of inhibitors were obtained for the 3 taphonomic processes (Table 2).
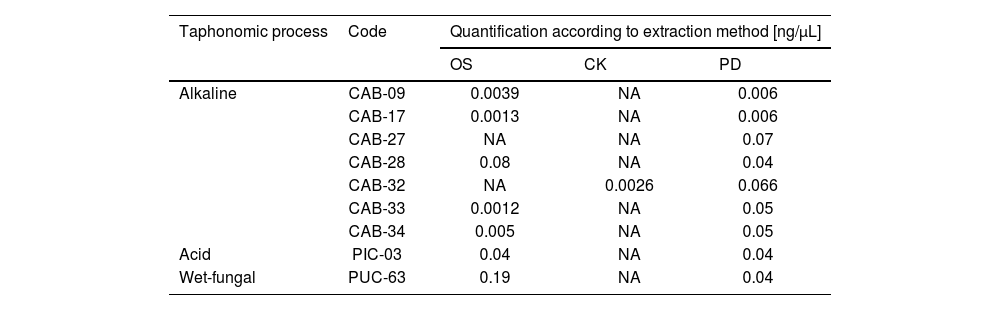
However, in 9 samples analysed (Table 3), there were discrepancies in terms of obtaining DNA for the 3 methods. According to these results, the extraction method using the commercial kit and the alkaline taphonomic process showed the highest number of samples with unsuccessful DNA extraction. The failure to obtain DNA was maintained even when silica columns were used as a second purification alternative for each case.
Samples in which there was a discrepancy in DNA retrieval efficiency.
| Taphonomic process | Code | Quantification according to extraction method [ng/μL] | ||
|---|---|---|---|---|
| OS | CK | PD | ||
| Alkaline | CAB-09 | 0.0039 | NA | 0.006 |
| CAB-17 | 0.0013 | NA | 0.006 | |
| CAB-27 | NA | NA | 0.07 | |
| CAB-28 | 0.08 | NA | 0.04 | |
| CAB-32 | NA | 0.0026 | 0.066 | |
| CAB-33 | 0.0012 | NA | 0.05 | |
| CAB-34 | 0.005 | NA | 0.05 | |
| Acid | PIC-03 | 0.04 | NA | 0.04 |
| Wet-fungal | PUC-63 | 0.19 | NA | 0.04 |
CK: commercial kit; PD: previous demineralisation; NA: not amplified; OS: organic solvent.
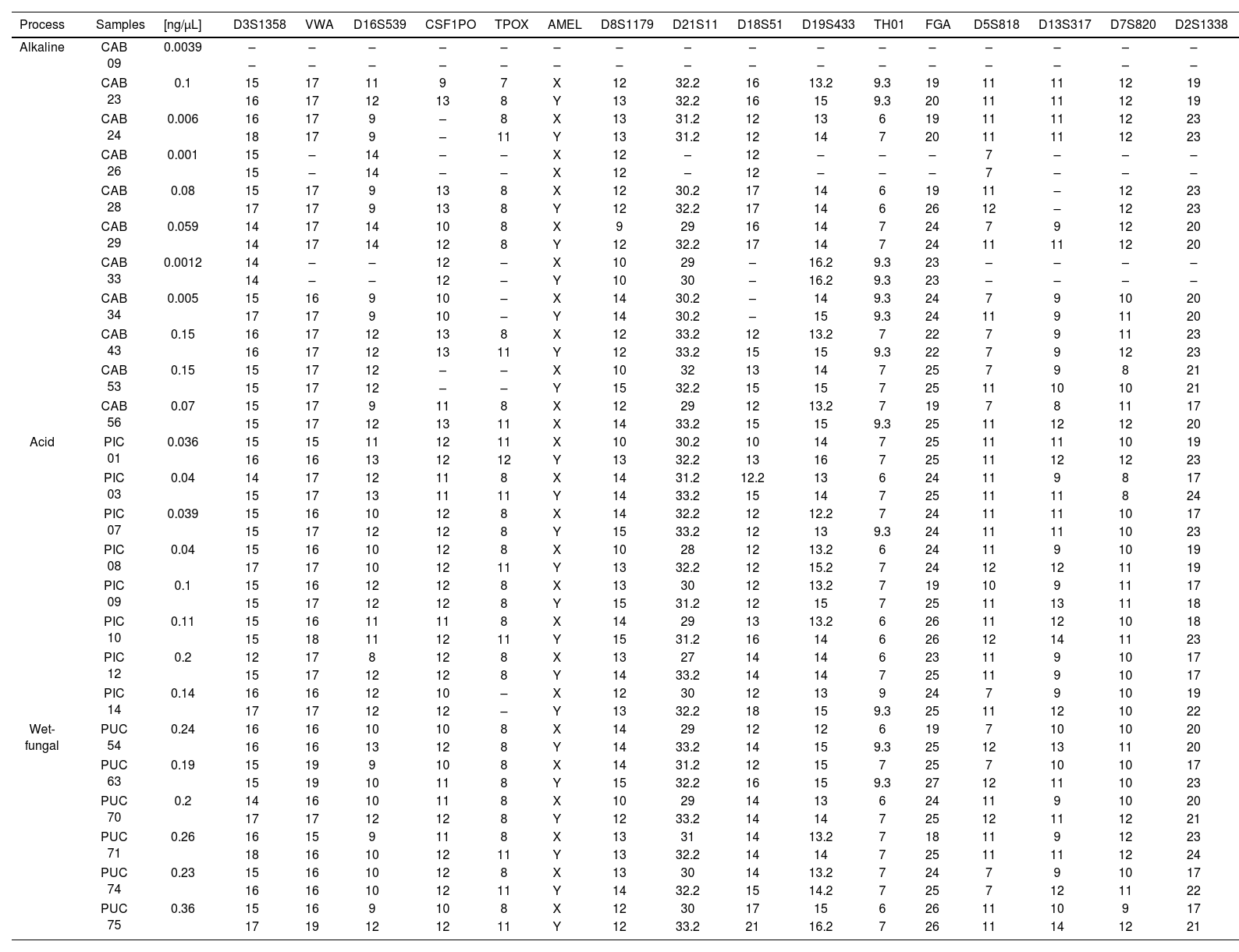
Table 4 shows the results of STR profiles obtained from DNA extracted by the organic solvent method. Identification showed that 99% of the samples corresponded to male remains, only 2 samples corresponded to a female source. The acid and wet-fungal processes were those in which 88% and 100% of the complete profiles were obtained, respectively. In contrast, the alkaline process had the most incomplete profiles, the CSF1PO and TPOX markers being the least identified. Finally, the D3S1358 and D8S1179 loci were loci that were identified for all samples.
STR obtained according to the taphonomic process.
| Process | Samples | [ng/μL] | D3S1358 | VWA | D16S539 | CSF1PO | TPOX | AMEL | D8S1179 | D21S11 | D18S51 | D19S433 | TH01 | FGA | D5S818 | D13S317 | D7S820 | D2S1338 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkaline | CAB 09 | 0.0039 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| CAB 23 | 0.1 | 15 | 17 | 11 | 9 | 7 | X | 12 | 32.2 | 16 | 13.2 | 9.3 | 19 | 11 | 11 | 12 | 19 | |
| 16 | 17 | 12 | 13 | 8 | Y | 13 | 32.2 | 16 | 15 | 9.3 | 20 | 11 | 11 | 12 | 19 | |||
| CAB 24 | 0.006 | 16 | 17 | 9 | – | 8 | X | 13 | 31.2 | 12 | 13 | 6 | 19 | 11 | 11 | 12 | 23 | |
| 18 | 17 | 9 | – | 11 | Y | 13 | 31.2 | 12 | 14 | 7 | 20 | 11 | 11 | 12 | 23 | |||
| CAB 26 | 0.001 | 15 | – | 14 | – | – | X | 12 | – | 12 | – | – | – | 7 | – | – | – | |
| 15 | – | 14 | – | – | X | 12 | – | 12 | – | – | – | 7 | – | – | – | |||
| CAB 28 | 0.08 | 15 | 17 | 9 | 13 | 8 | X | 12 | 30.2 | 17 | 14 | 6 | 19 | 11 | – | 12 | 23 | |
| 17 | 17 | 9 | 13 | 8 | Y | 12 | 32.2 | 17 | 14 | 6 | 26 | 12 | – | 12 | 23 | |||
| CAB 29 | 0.059 | 14 | 17 | 14 | 10 | 8 | X | 9 | 29 | 16 | 14 | 7 | 24 | 7 | 9 | 12 | 20 | |
| 14 | 17 | 14 | 12 | 8 | Y | 12 | 32.2 | 17 | 14 | 7 | 24 | 11 | 11 | 12 | 20 | |||
| CAB 33 | 0.0012 | 14 | – | – | 12 | – | X | 10 | 29 | – | 16.2 | 9.3 | 23 | – | – | – | – | |
| 14 | – | – | 12 | – | Y | 10 | 30 | – | 16.2 | 9.3 | 23 | – | – | – | – | |||
| CAB 34 | 0.005 | 15 | 16 | 9 | 10 | – | X | 14 | 30.2 | – | 14 | 9.3 | 24 | 7 | 9 | 10 | 20 | |
| 17 | 17 | 9 | 10 | – | Y | 14 | 30.2 | – | 15 | 9.3 | 24 | 11 | 9 | 11 | 20 | |||
| CAB 43 | 0.15 | 16 | 17 | 12 | 13 | 8 | X | 12 | 33.2 | 12 | 13.2 | 7 | 22 | 7 | 9 | 11 | 23 | |
| 16 | 17 | 12 | 13 | 11 | Y | 12 | 33.2 | 15 | 15 | 9.3 | 22 | 7 | 9 | 12 | 23 | |||
| CAB 53 | 0.15 | 15 | 17 | 12 | – | – | X | 10 | 32 | 13 | 14 | 7 | 25 | 7 | 9 | 8 | 21 | |
| 15 | 17 | 12 | – | – | Y | 15 | 32.2 | 15 | 15 | 7 | 25 | 11 | 10 | 10 | 21 | |||
| CAB 56 | 0.07 | 15 | 17 | 9 | 11 | 8 | X | 12 | 29 | 12 | 13.2 | 7 | 19 | 7 | 8 | 11 | 17 | |
| 15 | 17 | 12 | 13 | 11 | X | 14 | 33.2 | 15 | 15 | 9.3 | 25 | 11 | 12 | 12 | 20 | |||
| Acid | PIC 01 | 0.036 | 15 | 15 | 11 | 12 | 11 | X | 10 | 30.2 | 10 | 14 | 7 | 25 | 11 | 11 | 10 | 19 |
| 16 | 16 | 13 | 12 | 12 | Y | 13 | 32.2 | 13 | 16 | 7 | 25 | 11 | 12 | 12 | 23 | |||
| PIC 03 | 0.04 | 14 | 17 | 12 | 11 | 8 | X | 14 | 31.2 | 12.2 | 13 | 6 | 24 | 11 | 9 | 8 | 17 | |
| 15 | 17 | 13 | 11 | 11 | Y | 14 | 33.2 | 15 | 14 | 7 | 25 | 11 | 11 | 8 | 24 | |||
| PIC 07 | 0.039 | 15 | 16 | 10 | 12 | 8 | X | 14 | 32.2 | 12 | 12.2 | 7 | 24 | 11 | 11 | 10 | 17 | |
| 15 | 17 | 12 | 12 | 8 | Y | 15 | 33.2 | 12 | 13 | 9.3 | 24 | 11 | 11 | 10 | 23 | |||
| PIC 08 | 0.04 | 15 | 16 | 10 | 12 | 8 | X | 10 | 28 | 12 | 13.2 | 6 | 24 | 11 | 9 | 10 | 19 | |
| 17 | 17 | 10 | 12 | 11 | Y | 13 | 32.2 | 12 | 15.2 | 7 | 24 | 12 | 12 | 11 | 19 | |||
| PIC 09 | 0.1 | 15 | 16 | 12 | 12 | 8 | X | 13 | 30 | 12 | 13.2 | 7 | 19 | 10 | 9 | 11 | 17 | |
| 15 | 17 | 12 | 12 | 8 | Y | 15 | 31.2 | 12 | 15 | 7 | 25 | 11 | 13 | 11 | 18 | |||
| PIC 10 | 0.11 | 15 | 16 | 11 | 11 | 8 | X | 14 | 29 | 13 | 13.2 | 6 | 26 | 11 | 12 | 10 | 18 | |
| 15 | 18 | 11 | 12 | 11 | Y | 15 | 31.2 | 16 | 14 | 6 | 26 | 12 | 14 | 11 | 23 | |||
| PIC 12 | 0.2 | 12 | 17 | 8 | 12 | 8 | X | 13 | 27 | 14 | 14 | 6 | 23 | 11 | 9 | 10 | 17 | |
| 15 | 17 | 12 | 12 | 8 | Y | 14 | 33.2 | 14 | 14 | 7 | 25 | 11 | 9 | 10 | 17 | |||
| PIC 14 | 0.14 | 16 | 16 | 12 | 10 | – | X | 12 | 30 | 12 | 13 | 9 | 24 | 7 | 9 | 10 | 19 | |
| 17 | 17 | 12 | 12 | – | Y | 13 | 32.2 | 18 | 15 | 9.3 | 25 | 11 | 12 | 10 | 22 | |||
| Wet-fungal | PUC 54 | 0.24 | 16 | 16 | 10 | 10 | 8 | X | 14 | 29 | 12 | 12 | 6 | 19 | 7 | 10 | 10 | 20 |
| 16 | 16 | 13 | 12 | 8 | Y | 14 | 33.2 | 14 | 15 | 9.3 | 25 | 12 | 13 | 11 | 20 | |||
| PUC 63 | 0.19 | 15 | 19 | 9 | 10 | 8 | X | 14 | 31.2 | 12 | 15 | 7 | 25 | 7 | 10 | 10 | 17 | |
| 15 | 19 | 10 | 11 | 8 | Y | 15 | 32.2 | 16 | 15 | 9.3 | 27 | 12 | 11 | 10 | 23 | |||
| PUC 70 | 0.2 | 14 | 16 | 10 | 11 | 8 | X | 10 | 29 | 14 | 13 | 6 | 24 | 11 | 9 | 10 | 20 | |
| 17 | 17 | 12 | 12 | 8 | Y | 12 | 33.2 | 14 | 14 | 7 | 25 | 12 | 11 | 12 | 21 | |||
| PUC 71 | 0.26 | 16 | 15 | 9 | 11 | 8 | X | 13 | 31 | 14 | 13.2 | 7 | 18 | 11 | 9 | 12 | 23 | |
| 18 | 16 | 10 | 12 | 11 | Y | 13 | 32.2 | 14 | 14 | 7 | 25 | 11 | 11 | 12 | 24 | |||
| PUC 74 | 0.23 | 15 | 16 | 10 | 12 | 8 | X | 13 | 30 | 14 | 13.2 | 7 | 24 | 7 | 9 | 10 | 17 | |
| 16 | 16 | 10 | 12 | 11 | Y | 14 | 32.2 | 15 | 14.2 | 7 | 25 | 7 | 12 | 11 | 22 | |||
| PUC 75 | 0.36 | 15 | 16 | 9 | 10 | 8 | X | 12 | 30 | 17 | 15 | 6 | 26 | 11 | 10 | 9 | 17 | |
| 17 | 19 | 12 | 12 | 11 | Y | 12 | 33.2 | 21 | 16.2 | 7 | 26 | 11 | 14 | 12 | 21 |
(−) Not amplified.
Although the forensic field has always encountered many difficulties in obtaining genetic profiles from historical skeletal remains, the field of ancient DNA has shown the potential to increase the success rates of DNA retrieval from very old and degraded samples obtained from a variety of substrates and environments.12 This research study highlights the efficacy of 3 methodologies for DNA extraction and purification from bone samples around 25 years old, and is thus a source of information on the particularities of DNA preservation against deteriorating mechanisms (biotic and abiotic). In our study, we were able to determine that the methodologies of extraction with organic solvents and previous demineralisation were superior to the quantification results obtained with the extraction methodology used by the QIAgen® commercial kit in the 3 taphonomic processes.
With regard to the age of the samples and its relationship with yield in obtaining DNA, a recent study,13 using samples from 4 different periods (30, 800, 1500, and 2000 years), showed that the difference tends to be proportional to the age (averages of 15, 0.03, 0.4, and 0.2 pg/mg, respectively). However, at relatively short age times (from days to 62 years)14 and intrinsic sample age (27–66 years),15 the approaches demonstrate a greater difference in yield according to the extraction method employed, and could be an indication that, even at an age of 60 years, the yields may not be as meagre.
For our case, the approach of comparing DNA yields in relation to time correlates directly with the taphonomic processes: alkaline (29 years), acid (25 years), and wet-fungal (26 years). This peculiar association is due to the massive and simultaneous deaths of individuals (in cases of crimes against humanity). In this sense, the 29-year-old samples presented lower yields; however, the 26-year-old samples gave higher yields than the 25-year-old samples. This is possibly the result of the lesser exposure to the environment of the 26-year-old samples compared to the 25-year-old samples.
Methodologies for DNA extraction from bone remains are based on incubating the pulverised sample in an extraction buffer containing EDTA, which acts as a demineralising agent.11 As a result, the amount of residual calcium decreases progressively, therefore, as the mineral content is removed, the retained mass decreases with the thermal instability of the collagen. Thus, the demineralised bone matrix easily exposes the DNA molecule because it is free of mineral coating and collagen.16
The main problems encountered when working with bone samples include PCR inhibitors such as porphyrin residues, calcium carbonate deposits (CaCO3), manganese oxide (MnO2), and humic and fulvic acids, the latter belonging to the family of molecules that are abundant in soil (and make bone remains greyish, porous, and fragile).17 The presence of these compounds hinders the correct trajectory of the DNA through the membrane in the purification columns, which interferes with its amplification.18 Humic acid is postulated as the main inhibitor in the present methodology, because it is the major component of soil and affects the coupling of Taq polymerase at the elongation stage.19 Dithiothreitol (DTT) and alcohol, to name the most common, are other important inhibitors when working with bone remains.
Interestingly, the methodology using organic solvents records the highest number of samples with inhibitors and in turn the highest amount of retrieved DNA. A likely explanation for this is that the presence of salts is abundant in bone remains, which inverts the organic phase and the aqueous phase, leading to a generalised distribution of inhibitors that are usually in the organic phase.20 The literature indicates that to minimise this it is essential to use purification columns11 and centrifugation.21 This research corroborates the efficacy of the Amicon Ultra centrifugal filter units Ultra-4, MWCO 30 kDa, QIAamp, and Centricon® YM-100 columns in the removal of inhibitors.
Although there is no apparent correlation between taphonomic processes and the presence of inhibitors, the literature highlights that soils, according to their nature, tend to develop a certain vegetation layer and, mainly, generate the best conditions for the formation of humic acids and different physico-chemical processes that cause severe damage to the deposited bone structure,22 which would help to explain the existence of these types of inhibitory substances in acid and alkaline environments, primarily.23 Likewise, proteins and carbohydrates have been identified as the main contaminants in acidic and wet/fungal environments, while the alkaline environment would be subject to inhibitors due to the presence of carbohydrates alone.24
In terms of genetic profiling, the organic solvent-based extraction method gave a higher number of positive amplifications (complete genetic profiles). This method is the most widely used in forensic analysis, despite being a very laborious technique involving large volumes of toxic reagents.25 Its yield lies in prior protein digestion24 and the subsequent action of phenol and chloroform to denature the proteins and dissolve the lipids respectively, which allows the nucleic acids to be soluble in the aqueous phase, so that the difference in density between the aqueous and organic phases (phenol and chloroform) makes it possible to separate them by centrifugation.26
Finally, the DNA concentrations obtained in the present study are, for the most part, relatively low and are consistent with those obtained by other authors.14,27,28 However, as in the literature, it was possible to obtain genetic profiles, since current sequencing methodologies are increasingly sensitive in the detection of nucleic acids.29,30
ConclusionsAccording to our results, the methodology using organic solvents and previous demineralisation proved to be optimal for DNA extraction compared to the method using the commercial kit. Although the method using organic solvents presented a higher number of inhibitors, the higher concentration values obtained were used to obtain STR profiles after repurification. Finally, the alkaline taphonomic context proved the most drastic scenario for DNA degradation because most of the profiles were incomplete compared to the other 2 contexts.
The authors would like to thank the management and forensic analysts of the Unidad de Biología Molecular y Genética (UNBIMOG) del Instituto de Medicina Legal y Ciencias Forenses del Ministerio Público (Molecular Biology and Genetics Unit (UNBIMOG) of the Institute of Legal Medicine and Forensic Sciences of the Public Prosecutor's Office). This research study is part of the thesis of Lucero I. Portuguéz Ramírez for her master's degree in Genetics in the Postgraduate Programme of the Faculty of Biological Sciences of the Universidad Nacional Mayor de San Marcos.
Please cite this article as: Portuguéz Ramírez LI, Vivas-Ruiz DE, Parra Chinchilla RC, Rivera Fernández NO. Eficacia de 3 métodos de extracción de ADN a partir de restos óseos obtenidos de 3 contextos tafonómicos. Revista Española de Medicina Legal. 2023. https://doi.org/10.1016/j.reml.2022.11.004.