Hospitals are considered an excellent compartment for the selection of resistant and multi-drug resistant (MDR) bacteria. The overuse and misuse of antimicrobial agents are considered key points fuelling this situation. Antimicrobial stewardship programs have been designed for better use of these compounds to prevent the emergence of resistant microorganisms and to diminish the upward trend in resistance. Nevertheless, the relationship between antibiotic use and antimicrobial resistance is complex, and the desired objectives are difficult to reach. Various factors affecting this relationship have been advocated including, among others, antibiotic exposure and mutant selection windows, antimicrobial pharmacodynamics, the nature of the resistance (natural or acquired, including mutational and that associated with horizontal gene transfer) and the definition of resistance. Moreover, antimicrobial policies to promote better use of these drugs should be implemented not only in the hospital setting coupled with infection control programs, but also in the community, which should also include animal and environmental compartments. Within hospitals, the restriction of antimicrobials, cycling and mixing strategies and the use of combination therapies have been used to avoid resistance. Nevertheless, the results have not always been favorable and resistant bacteria have persisted despite the theoretical benefits of these strategies. Mathematical models as well as microbiological knowledge can explain this failure, which is mainly related to the current scenario involving MDR bacteria and overcoming the fitness associated with resistance. New antimicrobials, rapid diagnostic and antimicrobial susceptibility testing and biomarkers will be useful for future antimicrobial stewardship interventions.
Los hospitales son considerados un excelente compartimento para la selección de bacterias resistentes y multirresistentes. La propia utilización de antimicrobianos y su uso inadecuado se consideran factores esenciales que determinan esta situación. Los programas de mejora en el empleo de estos fármacos han sido diseñados para evitar la aparición de microorganismos resistentes e invertir la tendencia de aumento actual de la resistencia. No obstante, la relación entre el uso de antibióticos y la resistencia es compleja y los objetivos no son siempre fáciles de conseguir. Hay diferentes factores que afectan esta relación que incluyen, entre otros, la exposición al antimicrobiano y la ventana de selección, su farmacodinamia, la naturaleza de la resistencia (natural o adquirida, incluyendo la resistencia mutacional y la asociada a la transferencia de genes de resistencia) y la propia definición de resistencia. Asimismo, las políticas de uso de antimicrobianos para promover su mejor utilización deben ser implantadas no solo en el hospital, asociadas a programas de control de infección, sino también en la comunidad, que también debe incluir el compartimento animal y medioambiente. En el hospital se han empleado diferentes estrategias para evitar la resistencia, entre ellas la restricción de los antimicrobianos, su utilización en ciclos o en “mezcla” y las terapias combinadas. No obstante, los resultados no siempre han sido favorables y las bacterias han persistido a pesar de los beneficios teóricos de las estrategias implantadas. Los modelos matemáticos y el conocimiento microbiológico pueden explicar estos fracasos, que están esencialmente relacionados con el actual escenario de multirresistencia y la superación del coste inherente de la resistencia. Los nuevos antimicrobianos, el diagnóstico rápido, la mejora en las pruebas de determinación de sensibilidad y el uso de los biomarcadores serán útiles en la implantación de los programas de mejora del uso de antimicrobianos.
The increasing resistance rates to antimicrobial agents have been indefectibly associated with the use of these drugs. It is well known that the inappropriate use and overuse of these agents has accelerated this process over the last 80 years to the dramatic levels that have made scientists, health authorities and politicians aware of the problem.1–3 Stewardship programs have been designed for better use of antimicrobial agents not only to prevent the emergence of resistant bacteria, but also to diminish the upward trend in resistance.4 Current knowledge of microbiological aspects of resistance, such as the natural presence of resistance determinants in environmental bacteria, and the emergence and selection processes in pathogenic bacteria, have raised awareness in the scientific community.5–7In addition, information has accumulated on the relationship of pharmacokinetics and pharmacodynamics of antimicrobials in the selection processes in pathogenic bacteria and also in bacterial communities belonging to the normal human microbiome. This information has driven the modification of traditional dosing and duration schemes of antimicrobial treatments and has improved our understanding of the differences between microbiological resistance and clinical resistance.8,9 The objective of this article is to summarize the complex relationship of antibiotic use and antimicrobial resistance from a microbiological perspective in the light of various compartments (human, animal and environmental) and clinical evidence.
Microbiological factors influencing the relationship between antimicrobial use and resistanceWhile antimicrobial resistance can be regarded as an unavoidable consequence of the use of antibiotics, the interaction between these two elements is far from a simple linear equation. Indeed, each of the two variables involving the exposure of the microorganism to the antibiotic and the development of antibiotic resistance are extremely complex, as are their interactions.
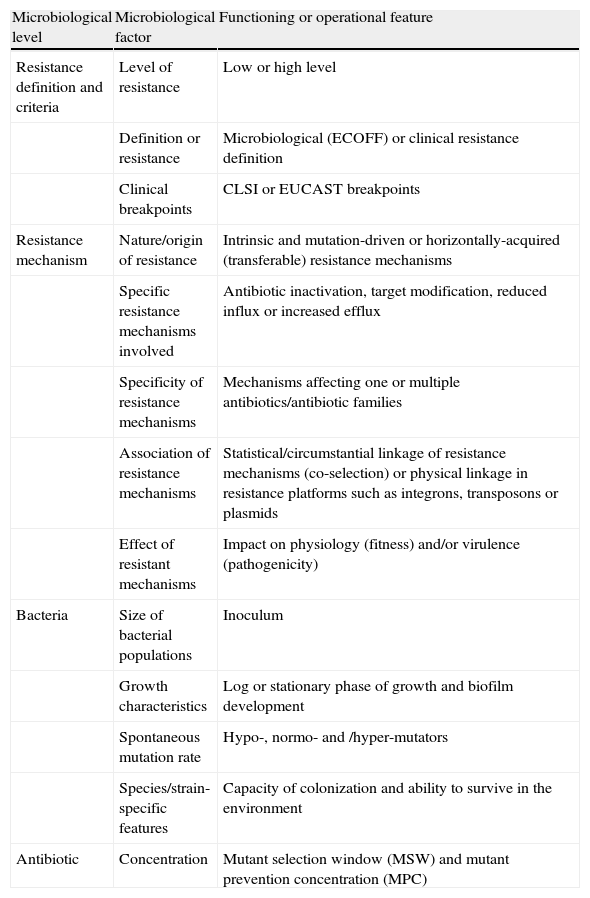
From the bacterial perspective, the emergence, enrichment and spread of antibiotic resistance is highly influenced by several factors that are summarized in Table 17,10.
Factors influencing the emergence, enrichment and spread of antibiotic resistance from a microbiological perspective.
| Microbiological level | Microbiological factor | Functioning or operational feature |
| Resistance definition and criteria | Level of resistance | Low or high level |
| Definition or resistance | Microbiological (ECOFF) or clinical resistance definition | |
| Clinical breakpoints | CLSI or EUCAST breakpoints | |
| Resistance mechanism | Nature/origin of resistance | Intrinsic and mutation-driven or horizontally-acquired (transferable) resistance mechanisms |
| Specific resistance mechanisms involved | Antibiotic inactivation, target modification, reduced influx or increased efflux | |
| Specificity of resistance mechanisms | Mechanisms affecting one or multiple antibiotics/antibiotic families | |
| Association of resistance mechanisms | Statistical/circumstantial linkage of resistance mechanisms (co-selection) or physical linkage in resistance platforms such as integrons, transposons or plasmids | |
| Effect of resistant mechanisms | Impact on physiology (fitness) and/or virulence (pathogenicity) | |
| Bacteria | Size of bacterial populations | Inoculum |
| Growth characteristics | Log or stationary phase of growth and biofilm development | |
| Spontaneous mutation rate | Hypo-, normo- and /hyper-mutators | |
| Species/strain-specific features | Capacity of colonization and ability to survive in the environment | |
| Antibiotic | Concentration | Mutant selection window (MSW) and mutant prevention concentration (MPC) |
CLSI: Clinical and Laboratory Standards Institute; ECOFF: epidemiological cut-off values; EUCAST: European Committee of Antimicrobial Susceptibility Testing.
A major issue in assessing the impact of antimicrobial use in resistance is the definition of resistance itself. There are basically two points of view to define resistance, based on clinical or microbiological approaches. Clinical susceptibility and resistance breakpoints are applied according to whether the in vitro inhibitory concentrations of the antibiotics are associated with a high probability of success or failure of antimicrobial therapy.11 There are essentially two official agencies establishing these clinical breakpoints: the Clinical Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility testing (EUCAST). The lack of harmonization of clinical breakpoints by these international agencies has a major impact on resistance definition and prevalence.12
Clinical breakpoints do not necessarily reflect the presence or absence of resistance mechanisms, adding further complexity to the equation. Indeed, from the microbiological point of view, resistance is defined as the presence of a resistance mechanism in a strain that reduces its susceptibility compared to that of wild-type strains within that species. Thus, microbiological breakpoints, defined as epidemiological cut-off (ECOFF) values by EUCAST, are based on the analysis of the distribution of MICs in wild-type strains.13 The use of ECOFF values in assessing the impact of antibiotic use in resistance widens the spectra of resistance mechanisms analyzed, because low-level resistance mechanisms are not always covered by clinical breakpoints.7
Microbiological breakpoints are particularly relevant for estimating the impact of low-level resistance mechanisms, which frequently preclude the emergence of clinical resistance.14 Still, this strategy is not entirely consistent with the direct phenotypic or genotypic assessment of resistance mechanisms, due to certain MIC overlapping between those strains harboring or not a given resistance mechanism. First step fluoroquinolone resistance mutations and extended spectrum β-lactamases (ESBL) or carbapenemase production in Enterobacteriaceae are classical examples in which the use of specific phenotypic tests improves the quantification of the resistance burden.15,16
Antibiotic exposure, PK/PD parameters and mutant selection windowThe variable exposure, rather than the total antibiotic consumption, is based on the concentrations over time of the antibiotic (pharmacokinetics, PK) producing an effect on a given microorganism over time (pharmacodynamics, PD) in a patient (or other host), and are known as PK/PD indices.17 For some antibiotics, such as aminoglycosides, the activity is known to be highly dependent on the maximum concentration reached, while for others, such as β-lactams, it is more dependent on the exposure time. Along these lines, three PK/PD parameters have been established to predict antimicrobial efficacy: the peak concentration divided by the minimum inhibitory concentration (MIC) (Cmax/MIC) for concentration dependent antibiotics, the time above the MIC (t > MIC) for time-dependent antibiotics and the area under the 24h concentration-time curve above MIC (AUC/MIC) for antibiotics with intermediate properties such as fluoroquinolones.18 Clearly, these parameters are useful for assessing the antimicrobial efficacy against susceptible populations, and are therefore increasingly incorporated into the rational for establishing clinical breakpoints, particularly by EUCAST.9
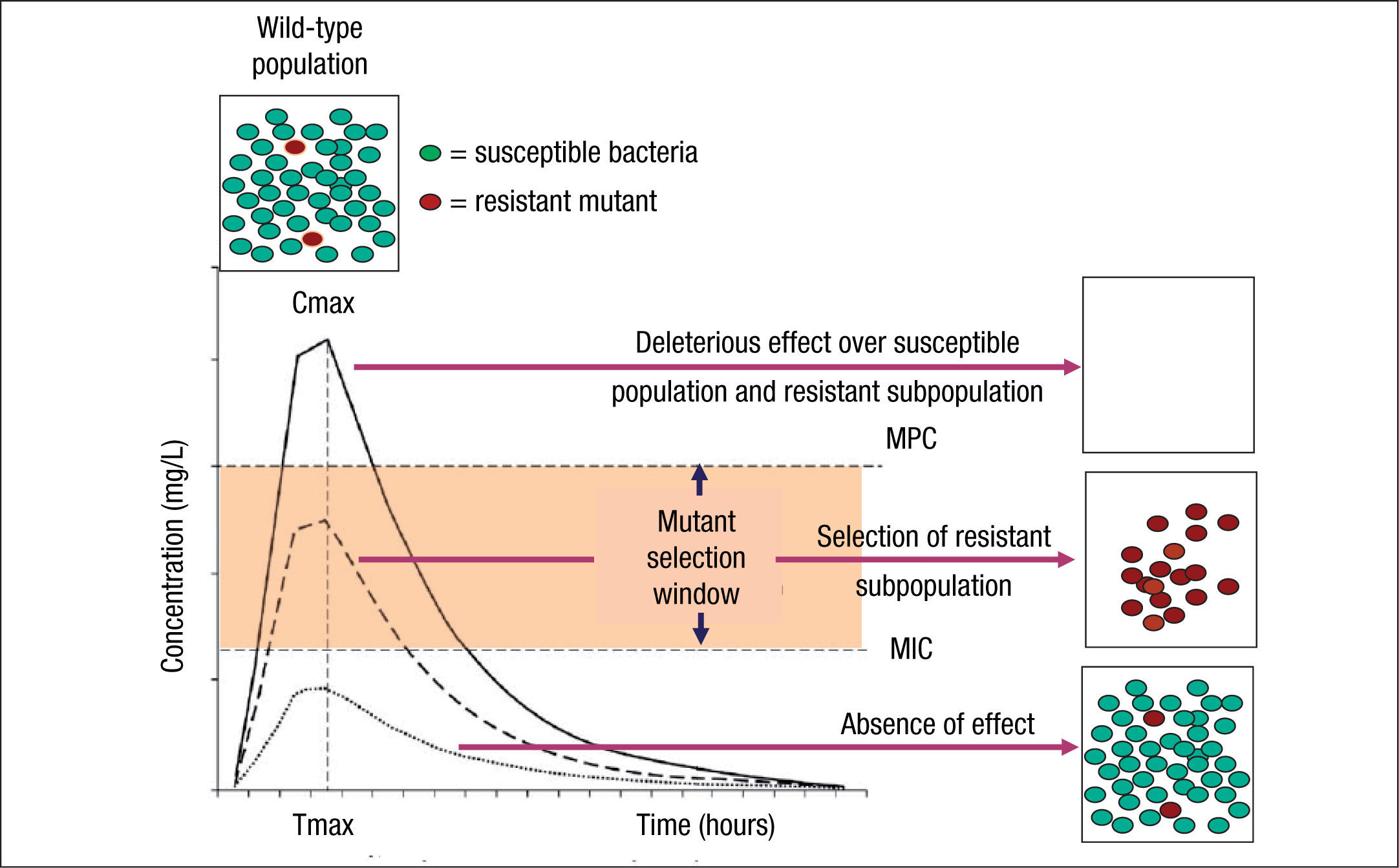
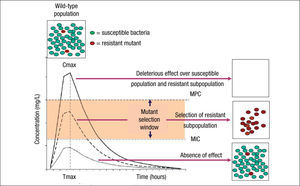
Although the use of PK/PD parameters increases the correct use of antimicrobial agents, understanding their significance in terms of clinical outcomes requires more research on some of the antimicrobial classes.19 In addition, the impact of these PK/PD parameters and derived breakpoints on antimicrobial resistance development is not yet well established. Indeed, the relationships between exposure intensity (e.g. AUC/MIC ratio) and efficacy and resistance development are completely different.8 The relationship between exposure intensity and efficacy is sigmoid and monotonic. That means that at very low values of exposure intensity there is no effect, whereas at larger values, the greater the exposure intensity, the greater the bactericidal effect up to a maximum value. On the other hand, the relationship between exposure and resistance selection is non-monotonic and has the shape of an inverted U.20 This inverted U is related to the mutant selection window (MSW) concept that implies that resistance occurs at concentrations between the MIC of wild-type cells and the MIC of the least susceptible one-step resistant mutants,21 which is defined as mutant prevention concentrations (MPC)22 (Fig. 1). Thus, in principle, antibiotic concentrations should be ideally maintained below the MIC or above the MPC to avoid resistance development. However, MPCs for several antibiotics and pathogens (particularly Pseudomonas aeruginosa) are frequently higher than those attainable at the infection site even for wild-type strains.7 It should also be noted that even sub-inhibitory concentrations may readily select resistant mutants, due to a higher fitness of these variants in the presence of the antibiotic.23 Moreover, sub-inhibitory concentrations of several antibiotics have been shown to induce increased mutation rates through the activation of the SOS system including error-prone polymerases, further contributing to resistance development.24
Mutant selection window (MSW) and mutant prevention concentration (MPC). Square boxes represent the bacterial population and curves the pharmacokinetics (concentration over time) of an antimicrobial agent. MSW is the concentration range in which resistant mutants can be selected and is delimited by the minimal inhibitory concentration (MIC) and the MPC. Above MPC, the selection of resistant mutant subpopulation should not be possible and the susceptible population is abolished, whereas below MIC values no effect over susceptible and resistant subpopulations is theoretically produced.
Resistant mutants are part of the normal susceptible population and emerge at a constant frequency. However, this frequency may vary depending on the antibiotic and the microorganism. When a susceptible bacterial population is exposed to an antimicrobial agent, the majority of the bacteria are killed or inhibited by the antimicrobial but the resistant mutant subpopulation may persist over time and may become dominant over the susceptible population. Persistence of antimicrobial use accelerates this process.6,25
Intrinsic and acquired resistanceIntrinsic resistance or innate resistance occurs when a resistance mechanism is characteristically present in the majority of the bacterial isolates for a species. As a result, the antimicrobial activity of the drug is insufficient, rendering it clinically useless. From a microbiological point of view, intrinsic resistance can be a result of: a) inherent difficulties for the antibiotic to reach its target due to impaired permeability of the bacterial envelope or the presence of drug exports systems (efflux pumps); b) the absence of antimicrobial targets or the presence of targets with low affinity; and c) the presence of mechanisms, such as enzymes, that inhibit or destroy the antibiotic. An example of intrinsic resistance is the resistance to carbapenems in Stenotrophomonas maltophilia due to the production of a chromosomal carbapenemase. Intrinsic resistance can be expressed not only with high but also with low-level MIC values close to the susceptible breakpoint.
In contrast to intrinsic resistance, acquired resistance by mutation or acquisition of resistant genes by horizontal genetic transfer (HGT) may be present or not in bacteria. Acquired resistance by gene mutation occurs spontaneously within a susceptible wild-type bacterial population at a very low frequency (<10–9). This small fraction of cells has mutations that impede antimicrobial action, allowing their selection during clinical treatment or antibiotic exposure. An example of acquired resistance by mutation is quinolone resistance due to mutations in chromosomal gyrA and/or parC genes. On the other hand, acquisition of resistant genes by HGT is an efficient mechanism that facilitates the rapid spread of antibiotic resistance genes among and between bacterial species. Elements responsible for the mobilization of bacterial genes included bacteriophages, plasmids and transposons/insertion sequences. The relative contribution of the three classes of elements to HGT varies between species. Nevertheless, in the majority of bacterial species with clinical relevance the best-characterized elements are plasmids and transposons/insertion sequences. Important resistance mechanisms among Gram-negative bacteria as ESBL or carbapenemases are codified in genes located in mobile genetic elements.
Targets and levels of selectionCo-resistance is defined as the presence of various resistance mechanisms encoded by mutated or acquired resistance genes.6 The consequence of this phenomenon is the generation of bacteria with multi-drug resistant (MDR) phenotypes affecting different antimicrobial classes. In environments with a high antibiotic pressure, such as the hospital setting, antimicrobials can act as selective forces for bacterial evolution. The use of antimicrobials can result in the selection of variants over the natural susceptible bacterial population with decreased antibiotic susceptibility due to mutation or acquisition of resistance genes by HGT. Moreover, mutations or resistance genes can accumulate in certain pathogenic bacterial lineages or clones, allowing them to survive in the presence of various antimicrobial compounds, increasing the opportunity for their spread. This complex phenomenon is named the selection and co-selection process. During these processes, a single antibiotic could select different MDR isolates and different antimicrobials could select an MDR isolate.
The co-selection process can occur at several hierarchical levels, with genes, genetic platforms harboring resistance genes and clones as the units of antimicrobial resistance for selection. Resistance genes, including both mutated and acquired genes, constitute the first step in bacterial selection. Antibiotic traits are naturally present in environmental bacteria involved in physiological or metabolic functions, but these traits express resistance in the presence of antibiotics (intrinsic resistance). In this sense, intrinsic resistance may be a consequence of global bacterial physiology. For example, efflux pumps present in several bacterial species such as P. aeruginosa play a role in detoxification of metabolic products, but can also affect other substances such as antimicrobials. All these natural “resistant genes” are called resistome, which constitute a natural reservoir of antibiotic resistance in the environment.26 These intrinsic resistant determinants can be mobilized and captured in genetic platforms that are able to be inserted into the genome of the human or animal bacterial pathogens, leading to resistant phenotypes (the acquisition of resistant genes). Genetic platforms related to the mobilization, insertion and expression of resistant genes (insertion sequences, integrons, transposon and plasmids) by HGT play an important role in the maintenance of antibiotic resistance, constituting the second level of selection. For example, Class-1 integrons are genetic structures found in Gram-negative organisms and are able to integrate several resistance genes such as those encoding carbapenemases and aminoglycoside modifying enzymes. The dissemination of this trait among bacteria allows the co-selection of carbapenem- and aminoglycoside-resistant isolates under antibiotic pressure.27
Finally, bacterial clones are the supra-level of selection. They are specific variants or clones within a bacterial population that are able to accumulate resistant genes by mutation or acquisition by HGT. These subpopulations are called “high-risk clones” and are defined as clones with an enhanced ability to colonize, spread, and persist in a variety of niches, with acquired adaptive traits that increase pathogenicity or antibiotic resistance.28 They constitute the main vehicles dispersing antibiotic resistance on a global scale.29,30 Currently, the extensive use of Multilocus Sequence Typing (MLST), a molecular epidemiological tool based on sequencing internal short regions (around 400-500 bp) of seven housekeeping genes, has facilitated the identification of these clones. In addition, genome-wide analyses have revealed important differences in the genetic repertoire of isolates belonging to high-risk clones when compared with susceptible isolates, which do not cluster in any high-risk clones.31 This distinct genetic repertoire includes, among others, resistant and virulence traits, insertion sequence elements, genomic islands etc. Currently, it is considered that these determinants may be adaptive elements that have improved the adaptation of this subpopulation to the environment. Within this landscape, antibiotics act as selectors and amplifiers of these clones, and antibiotic policies might restrict dispersion of these clones.32 Examples of these high-risk clones are Escherichia coli-ST131 or Klebsiella pneumoniae-ST258 associated with the global dissemination of blaCTX-M-15 and blaKPC respectively.30
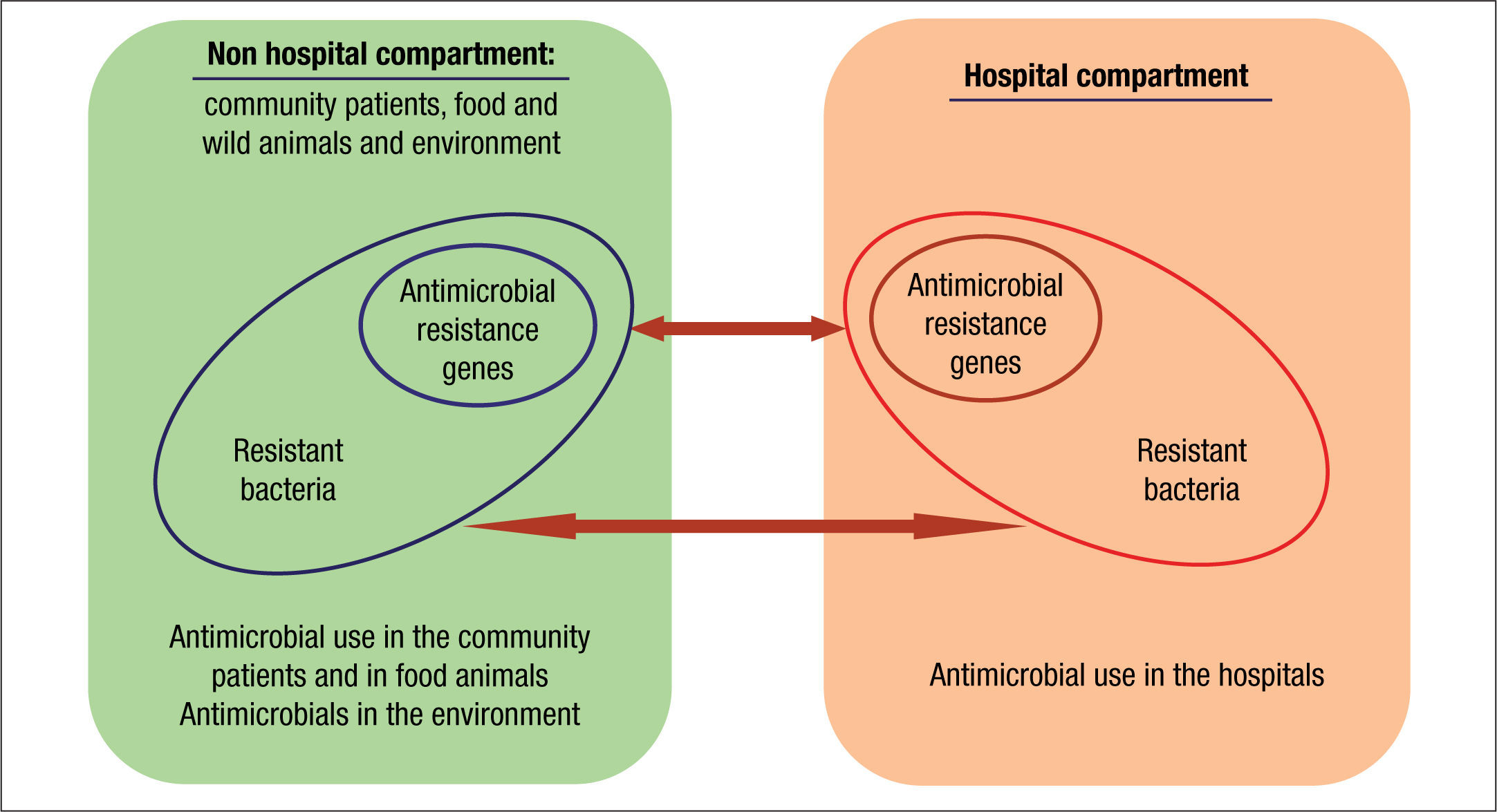
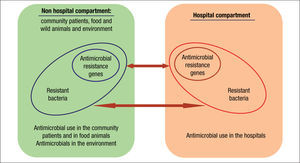
Compartments of selectionThe current organization of healthcare has almost blurred the traditional frontiers of the hospital compartment and the community. As a consequence, antibiotic policies and stewardship programs should be designed as a whole. These strategies should also include animal and environmental compartments due to their inter-connectivity (Fig. 2).33
Traditionally, hospitals have been considered the main reservoir of antimicrobial-resistant bacteria. The intestinal tract is the main reservoir of resistant bacteria among hospitalized patients. As previously stated, a recognized collateral effect of antimicrobial therapy is the possible selection of resistant microbiota. This altered microbiota may contaminate the hospital environment and from there it reaches other patients and surfaces via cross contamination. Around 20%-40% of nosocomial infections can be mainly attributed to cross-infection via the hands of healthcare personnel.34 Less frequently, patients can become colonized with nosocomial pathogens by direct contact with contaminated surfaces or equipment. In some cases, the extent of patient-to-patient transmission has been found to be directly proportional to the level of environmental contamination. Moreover, inadequate environmental cleaning and hygiene help maintain these microorganisms in the environment.
Some studies have identified the previous presence of a colonized or infected patient in a room as a risk factor for the acquisition of the same pathogen by a new occupant, presumably because of residual room contamination that is not removed through terminal cleaning and disinfection. This effect has been shown for vancomycin resistant enterococci (VRE), methicillin resistant S. aureus (MRSA), Clostridium difficile, MDR P. aeruginosa and Acinetobacter baumannii.35 Moreover, clean hands touching contaminated surfaces of equipment clearly diminish hand hygiene efficacy.36,37
It is well known that contaminating pathogens may survive for prolonged periods of time on inanimate surfaces with relative resistance to disinfectants.34 The persistence of nosocomial pathogens on inanimate surfaces was analyzed in well-written review.38 Most of them survive for months on dry surfaces but certain pathogens, such as P. aeruginosa and A. baumannii, need humidity for survival. This review emphasizes that these pathogens easily survive and transmit if surface disinfection is not performed. A recent study showed several-weeks survival of MDR E. coli and K. pneumoniae on stainless steel surfaces. More interestingly, the researchers also demonstrated that plasmid-mediated HGT of β-lactamase genes occurred when the donor and recipient cells were mixed on stainless steel. This study demonstrates that the ability of pathogens to persist in the environment, particularly on touch surfaces, may play an important role in HGT of resistance determinants.39 In addition, it has also recently been reported that the biofilm-producing A. baumannii strains survive on inanimate surfaces much longer than their non-biofilm-producing counterparts.40 In this scenario, inappropriate and excessive use of antimicrobials contributes to the persistence of resistant bacteria.41,42 Also, inadequate infection control helps to propagate these resistant bacteria, and may lead to outbreaks and endemics. Moreover, antibiotic pressure also converts hospital waste into a potential source of antimicrobial resistance in the environment outside the hospital. Antibiotics used in hospitals are mainly released non-metabolized into the aquatic environment via wastewater, contributing to an increase of the environmental resistance rates.43,44
Taking into account these facts together with the fact that antimicrobial use promotes the selection of MDR microorganisms, the hospital environment should be considered an important source of these pathogens. Consequently, programs to optimize antimicrobial use should be implemented in parallel with epidemiological measures.
Selection density and colonization pressureRegarding antimicrobial pressure in the hospital, it is important to consider the influence of “selection density”, which is the quantity of antibiotic used per individual in a geographic area.45 In the hospital, the selection density is higher when the numbers of antibiotics in the formulary are small and if there is no diversification strategy. Also the circulation of resistant clones and the accumulation of a few patients in small spaces such as the ICU or specific wards contribute to a higher selection density.
Patients’ microbiota is the source of bacteria that contaminates the hospital environment. Several studies have demonstrated a relationship between antimicrobial consumption and isolation of antimicrobial resistant bacteria.41,42 One classical study demonstrated that the amount of environmental antimicrobial resistant S. aureus and Gram-negative bacilli in the ICU correlated with the defined daily dose (DDD) of antibiotics used in that unit.46
The transmission of microorganisms is influenced by various factors in the hospital compartment. One of them is “colonization pressure” which is the proportion of patients in a given unit who are colonized during a defined period of time. Several studies have demonstrated that high colonization pressure increases the spread and transmission of MDR microorganisms such as MRSA, VRE and A. baumannii in hospitals.47,48 Therefore, additional infection-control measures should be implemented immediately after identification of the first carriers, rather than waiting for further dissemination of the pathogen, especially in the ICU or in other special units (immunocompromised, transplant recipients, etc.).
Community and animal compartmentsClassically, nosocomial pathogens were considered the source of drug resistant pathogens that were detected in the community. Nevertheless, there is growing evidence on the importance of the community as the origin of MDR pathogens that are introduced into the hospital setting. But where do they come from? Commensal bacteria of the intestinal or cutaneous microbiota of animals are also an important reservoir of resistance mechanisms that can reach humans in various ways. Industrial animal food production is the ideal setting for the selection and sharing of resistance genes. The use of antimicrobials as feed additives often results in sub-therapeutic dosing to animals bred in unhygienic conditions, an optimal scenario for resistance selection. Moreover, incomplete or mostly nonexistent waste treatment leads to the dissemination of resistance into the environment and humans.43,49
Antimicrobials are administered to food production animals for various purposes, including treatment of infected animals, prophylaxis to prevent infections and growth promotion. Estimates suggest that antibiotic use in animal feed accounts for 60-80% of total antimicrobial production in the United States and in the European Union. This use has played a major role in the development and spread of VRE and MDR Salmonella.50,51 Moreover, there are many reports worldwide on the presence of resistant bacteria in consumer meat products, including poultry, beef, and pork.51–53 In a study from Seville (Spain) and Pittsburg (USA), a high proportion of ESBL producing E. coli was detected in retail meat from both countries.54 Moreover, the concept that urinary tract infections (UTI) could have a zoonotic origin has recently emerged from a study showing a clonal link between E. coli from meat and UTI isolates from humans.55
Some authors have reported the emergence of a genetic lineage of MRSA called ST398, initially associated with farm animals,49 which is being detected with some concern in samples of animal foods (especially meat and milk). Furthermore, this clone is frequently detected in humans in contact with farm animals that may transmit it to the community. Using hospital surveillance data, one study found that occupational exposure to farm animals or proximity to farms was a risk factor for MRSA infection.56
Animal companions are another possible reservoir of MDR bacteria. In a recent study ESBL-producing Enterobacteriaceae were present in healthy cats and dogs, particularly those with a history of previous antibiotic treatment.57
Interventions against the development of antibiotic resistance in the community before it can enter in the hospitals are urgently needed. These strategies should stress specific antibiotic stewardship programs for this compartment.
Environmental compartment, MDR bacteria and antibiotic persistenceBacteria are one of the most important groups of organisms in soil and in other environmental compartments as well as in natural sewage. In soil, natural antibiotics from bacteria and fungi control the dynamics of bacterial populations by acting as signaling molecules.58 Nevertheless, antibiotics could also favor the emergence of resistant bacteria. This fact normally depends on the antibiotic concentration and its bioavailability. Fortunately, as most natural components are biodegradable, the selection of resistant flora is thought to be less frequent in natural environments. In contrast, most of the antimicrobial compounds used currently in human and veterinary medicine are synthetic or semi-synthetic compounds and they are often much more stable and not as biodegradable as natural antibiotics. As a result, they may persist in the environment for a long period of time. Furthermore, they often have a broader spectrum with a wider impact on bacterial populations.
The environment constitutes a resistance reservoir with organisms able to share resistance genes,26,59 and either resistance genes or resistant bacteria may invade the human compartment. The transfer as well as the emergence of new combinations of resistance genes will happen most frequently in compartments with high bacterial density, e.g., biofilms, which are present in most natural compartments such as sediments and soils. Waste disposal is the major source of antimicrobial-resistant pathogens entering in the environment.43 Many antimicrobials used in animal food production are poorly absorbed in the gut of the animals, and as much as 90% of the main compound is excreted in urine and up to 75% in feces.60 Animal waste is typically applied to land, generally onsite or within some miles of the farm. These practices contaminate air, water, and soils near both storage and field application sites.61 Waste contributes to the emergence and spread of resistant bacteria for the following reasons: a) resistant pathogens associated with infectious diseases are present in animal waste; b) waste contains stable antimicrobial agents; and c) HGT or resistant determinants occur within the waste environment. Also, the geographic concentration of large-scale agricultural operations has been associated with contamination of soils and irrigation water for food crops and with the presence of antimicrobial-resistant bacteria in vegetables.62
Another phenomena of concern is that most of the compounds used in medicine are only partially metabolized by patients and are then discharged into the hospital or community sewage system. Along with excreta, they flow with municipal wastewater to the sewage treatment plant. They may pass through the sewage system and end up in the environment, mainly in the water compartment. A striking recent study demonstrated that exchange is taking place between the ESBL E. coli populations in infected humans and sewage sludge, most likely by the entry of ESBL E. coli from UTIs into the sewage system.63
The expected and the unexpected impact of antibiotic use on resistanceVarious general conclusions can be established when a review of the literature on antibiotic use and impact on resistance is performed: a) there is a direct correlation between specific antimicrobial use and antimicrobial resistance: an increase in antimicrobial use generally increases antibiotic resistant bacteria and a decrease in antimicrobial use decreases, at a certain level, antibiotic resistant bacteria; b) higher resistance levels are found in bacteria recovered from scenarios with high antibiotic density; best exemplified in the ICUs; c) patients with infections due to resistant and MDR organisms have been treated with more antimicrobials than those infected with susceptible organisms (the antimicrobial use is a risk factor for resistance); d) antimicrobial use increases the risk of selection of resistant bacteria in the normal microbiota; and e) prolonged antimicrobial use increases the risk of an infection due to resistant or MDR organisms. Although all these correlations seem obvious, the results expected when applying antibiotic policies strategies are not always obtained.64
Restrictions on antimicrobial useMost of the studies that correlate antibiotic use and resistance have been performed using specific microorganisms as indicators such us MRSA, VRE, ESBL or carbapenemase producing Enterobacteriaceae and MDR P. aeruginosa or A. baumannii. However, few studies have evaluated the impact on patients’ microbiota, known as the collateral effect, and few have considered the impact on the resistant pool in specific compartments (e.g., the hospital resistome).65
On the other hand, the number of studies addressing the decrease of antibiotic use and the impact on the decrease of resistance is much lower than those addressing the opposite; the increment of antibiotic use and the increase of resistance. The reasons are in most cases related to the absence of a rapid noticeable effect, failure of decreasing resistance rates and that a “time-series analysis” is needed to observe desirable results. Again, this can be partially explained from a microbiological perspective. The absence of the use of a specific antibiotic is normally associated with the increased use of another antibiotic that normally impacts the same bacteria due to its MDR nature, exemplifying the co-selection processes.6 This result has been observed not only in the hospital setting but also in the community. In an interesting experience of restriction of the use of sulfonamides in The United Kingdom and Sweden, the decrease of these antibiotics over years was not followed by a significant decrease of sulfonamide resistance in E. coli. This result was explained by the presence of sulfonamide resistance genes on the integron platforms that were not cleared despite the reduction in selection processes.66,67 This study demonstrated that genetic platforms recruiting resistant determinants undoubtedly hinder the clearance of resistant organisms.
However, the potential decrease in specific, resistant bacteria can be accompanied by an increase in another one. This fact was coined the “sequencing the resistance balloon”.68 In the late nineties, the emergence of imipenem resistant A. baumannii in various institutions was associated with the decrease of ceftazidime use due to the emergence of ESBL-producing K. pneumoniae, but with a parallel increase of imipenem prescriptions to treat infections due to this organism. More recently, the restriction of extended spectrum cephalosporin use has not always been associated with the decrease of ESBL-producing Enterobacteriaceae, probably due to the increasing use of fluoroquinolones. These compounds are risk factors for infections due to ESBL-producing bacteria. Moreover, they have been associated with the increasing rates of Enterobacteriaceae with mutations in the QDRD region of gyrA and with the spread of plasmid mediated quinolone resistant determinants (PMQR) such as qnr, aac(6’)-Ib-cr of qep genes, all frequently found resistant mechanisms in ESBL-producing Enterobacteriaceae.69,70 Unexpectedly, and despite worldwide increases in fluoroquinolone use, increasing resistance rates have not been observed in other bacteria, such as S. pneumoniae. This increase is probably due to the fitness cost when fluoroquinolone resistance emerges in this organism.71
Antimicrobial use, fitness cost and mathematical modelsOverall, the evidence suggests that in some cases bacterial resistance produces a fitness cost. In the absence of the antibiotic, it can be assumed that the resistant phenotype will disappear over time. If no cost of resistance is generated, it is likely that the resistant phenotype will persist although antibiotic pressure decreases or disappears.
Based on the assumptions above, it is possible to mathematically model the dynamics of development of resistance according to antibiotic consumption. These models indicate that the frequency of resistance will increase rapidly if the use of the antibiotic in question (proportion of individuals treated in a given community) exceeds a certain threshold, the magnitude of which is inversely related to the fitness of the resistant bacteria and directly related to the horizontal transmission rate.72 Once the threshold is exceeded, a constant resistance prevalence will be reached, the level of which will depend on the degree to which antibiotic consumption exceeds the threshold. The models also demonstrate that the decline in the prevalence of resistance in stable human communities (with low turnover population) associated with a decrease or cessation of antibiotic pressure will take much longer (typically several years) than was taken to generate it. Therefore the sooner measures are taken the better. Some of these models have been fitted to high turnover populations, such as hospitalized patients from the community, assuming that the number of admitted individuals who are colonized or infected with the microorganism of interest (e.g. MRSA or β-lactam-resistant P. aeruginosa) is very low. In these circumstances, the forecast models are similar, but the time required for the generation of resistance and, especially, for the decrease of its prevalence in response to the reduction of the antibiotic consumption, is counted in weeks rather than in years, although the fitness of both phenotypes is the same. An interesting aspect of nosocomial models is that the introduction of a second antibiotic capable of removing both the susceptible and resistant phenotypes to a given antibiotic should lower the prevalence of resistance to the latter antibiotic, suggesting that the concurrent use of antibiotics with various mechanisms of resistance in a community may not only not be deleterious, but may also be beneficial for decreasing the prevalence of resistance associated with the use of each.
Cycling and mixing strategiesVarious proposals have been performed and applied to decrease the selection process and control the evolution of antimicrobial resistance. One approach was antibiotic cycling (or rotation), which became very popular in ICUs in the early 2000 but has now been abandoned.73 The practice of antibiotic cycling involves a pattern of using defined, equal periods of time for the introduction and removal of an antibiotic (or an antibiotic class) possessing similar spectrum of activity by preferentially one of different mechanisms of action and resistance as the predominant choice. This is followed by a different group of antibiotics and rotated back through the chosen list of agents (e.g., the rotation of ceftazidime, piperacillin/tazobactam, cefepime and ciprofloxacin). The period of rotation is predefined on a scheduled basis. From a theoretical point of view, cycling attempts to minimize selection pressure. Nevertheless, the mathematical model and clinical and microbiological follow-up of resistance rates showed that cycling should no longer be recommended.74–76 Genetic and surveillance studies suggest that microorganisms were able to persist and acquire various resistance mechanisms with the increasing prevalence of MDR isolates.6
An alternative to cycling is the mixing (or heterogeneity) strategy. In this strategy a free selection of antibiotics by the clinician is allowed, according to patient's characteristics, personal preference, knowledge and experience. Selection density is lower, thus is the potential emergence of resistance. This was demonstrated in a prospective study performed in a single ICU with four sequentially implemented empirical antibiotic strategies for ventilator-associated pneumonia that included rotation and mixing strategies.77 Benefits of cycling were few when compared with mixing strategies. It is interesting to note that the superiority of mixing increases when implemented with specialist supervision. This strategy is known as PAMS (periodic antibiotic monitoring and supervision). Reductions in resistance rates were significant for mechanisms involving mutations and to a lesser extent or inexistent with acquired resistance mechanisms (e.g., ESBL-producing Enterobacteriaceae).78 Co-selection processes may be responsible for this effect.6
Combination therapyThe results of combination therapy to prevent the development of antimicrobial resistance are conflicting.79 Despite clear in vitro advantages, clinical studies have not conclusively confirmed reductions in resistance with the use of combination therapy. This result may be due to inadequate combination from a pharmacological point of view (e.g., low accessibility of one of the antimicrobials to the infection site, reducing potential benefits, or the preexistence of MDR bacteria that can be selected by all the antimicrobials used in the combination. This could be the case with β-lactam and aminoglycoside combinations in pulmonary infections, as aminoglycosides reach the respiratory tract inefficiently. Moreover, MDR bacteria are commonly resistant to antibiotics currently used in combination, thus avoiding the elimination of these bacteria due to co-selection.6
Future perspectives: new antimicrobials, rapid diagnostic and antimicrobial susceptibility testing, biomarkers and study designThe use of new antimicrobials is expected to have a favorable impact on resistance development. Future antimicrobial interventions will include targeted antimicrobials rather than broad-spectrum agents having few or no selection processes.80 This should be coupled with research on new antimicrobial targets with compounds associated with low frequencies of resistance development. Also, as new antimicrobials will have a narrower spectrum of activity, their use should be implemented with rapid bacterial identification and antimicrobial susceptibility testing.81 This approach will facilitate an early response and appropriate antibiotic choice.
On the other hand, the use of biomarkers will also be useful to define not only when an antibiotic is needed but also when the antimicrobial treatment should be stopped.82 The selection impact on the infective microorganisms and on the microbiota (the collateral effect) will be lower. These strategies will need to be implemented with careful monitoring and microbiological follow-up of resistant and MDR organisms. Moreover, the design of future studies addressing antibiotic use and antibiotic resistance should take into consideration the complexity of this association.83
FundingThis review was supported in part by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III -co-financed by the European Development Regional Fund “A way to achieve Europe” ERDF, the Spanish Network for the Research in Infectious Diseases (REIPI RD12/0015). Patricia Ruiz Garbajosa is partially supported by a research contract from the European Commission (R-GNOSIS-FP7-HEALTH-F3-2011-282512).
Conflicts of interestThe authors declare that they have no conflicts of interest.