To study the resistance of biofilms developed by non-pigmented rapidly growing mycobacteria (NPRGM) against amikacin, ciprofloxacin and clarithromycin in an in vitro model using clinical strains of different species.
DesignAntimicrobial susceptibilities of different clinical strains of Mycobacterium abscessus, Mycobacterium chelonae, Mycobacterium fortuitum, Mycobacterium peregrinum, Mycobacterium mucogenicum and Mycobacterium mageritense using conventional techniques were measured. Biofilm resistance was measured by using the sandwich technique developed by Anderl et al. using a concentration of antibiotic of 50mg/L. Penetration of antibiotics through biofilm was measured using the same technique with minimal modifications.
ResultsNPRGM biofilms showed drug resistance (percentages of viable bacteria >1% of those of controls) against antibiotics that are commonly used for the treatment of infections caused by these organisms, although there are intraspecies differences between strains. We have detected differences in antibiotic penetration through biofilms with an important permeability barrier for ciprofloxacin. However, other mechanisms must be probably more important to explain the antimicrobial resistance of NPRGM biofilm.
ConclusionsBiofilms formed by NPRGM are resistant to amikacin, ciprofloxacin and clarithromycin. As no resistance differences between the tested antibiotics have been observed, it is likely that biofilm permeability of antibiotics is of low importance for antimicrobial resistance of biofilms.
Estudiar la resistencia de biopelículas formadas por micobacterias no pigmentadas de crecimiento rápido (MNPCR) frente amicacina, ciprofloxacino y claritromicina en in modelo in vitro empleando aislamientos clínicos de diferentes especies.
Material y MétodosSe estudiaron las sensibilidades de las diferentes cepas clínicas de Mycobacterium abscessus, Mycobacterium chelonae, Mycobacterium fortuitum, Mycobacterium peregrinum, Mycobacterium mucogenicum y Mycobacterium mageritense mediante técnicas convencionales. La resistencia de dichas bacterias en la biopelícula fue estudiada mediante la técnica de sándwich descrita por Anderl et al. utilizando una concentración de antibiótico de 50mg/L. La penetración de los antibióticos a través de la biopelícula fue estudiada mediante la misma técnica con mínimas modificaciones.
ResultadosLas biopelículas de MNPCR presentaron resistencia (porcentajes de bacterias viables > 1% de los recuentos obtenidos en los controles) frente a todos los antibióticos que son empleados habitualmente en las infecciones causadas por estos organismos, si bien se detectaron diferencias dentro de la misma especie entre las diferentes cepas. Hemos detectado diferencias en la penetración de antibióticos a través de la biopelícula, especialmente con una importante disminución de la permeabilidad frente a ciprofloxacino. Sin embargo, otros mecanismos son, probablemente, más importantes para explicar la resistencia antimicrobiana de las biopelículas de MNPCR.
ConclusionesLas biopelículas formadas por MNPCR son resistentes frente a amicacina, ciprofloxacino y claritromicina. Como no se demostraron diferencias importantes entre los distintos antibióticos, es probable que la permeabilidad de la biopelícula frente a los antibióticos tenga poca importancia en la resistencia antimicrobiana de las biopelículas.
Non-pigmented rapidly growing mycobacteria (NPRGM) include different species of mycobacteria with common phenotypical characteristics1 and some of them,, such as Mycobacterium abscessus, Mycobacterium fortuitum or Mycobacterium chelonae, have been known to be human pathogens for a long time.All the species involved in this group are environmental organisms that became human pathogens under special circumstances or risk factors.2 Different studies have recovered NPRGM in biofilms associated with water systems,3 and a recent study has shown that most human NPRGM infections are biofilm-related.4 These infections include a broad spectrum of clinical syndromes, such as catheter-related bloodstream infections,5 surgical site infections,6 prosthetic joint-related infections,7–14 respiratory tract infections in cystic fibrosis patients12–14 and others.1,2,4In vitro studies have also demonstrated biofilm formation by almost all NPRGM, both in clinical and collection isolates, and a relationship between biofilm development ability and the production of human clinical disease.15,16
Biofilm development is an important factor in antimicrobial resistance. In this sense, different studies have shown the in vitro resistance of biofilms formed by NPRGM against disinfectants17 or antibiotics,18 including amikacin and clarithromycin resistance of M. abscessus embedded in biofilm.19
However, despite these observations, only a few reports have tried to determine the mechanisms of antimicrobial resistance among clinically relevant species of NPRGM. In this study, we have evaluated the permeability of different antimicrobials to cross over the biofilms developed by 6 different NPRGM species, as one of the factors that could be involved in the increased drug resistance of the biofilm.
Material and methodsStrainsTwenty biofilm-producing NPRGM clinical strains belonging to six different species (2M. abscessus, 2M. chelonae, 4M. fortuitum, 3M. peregrinum, 1M. mucogenicum and 2M. mageritense) were analyzed. These species were selected because they are the ones most commonly isolated NPRGM in human samples. These strains were identified by using a broad battery of biochemical tests (nitrate reduction, urease, 3-day arylsulphatase, Tween 80 hydrolysis, growth on McConkey agar without crystal violet, and use of citrate, mannitol, inositol, sorbitol and rhamnose) and PCR-restriction enzyme analysis (PRA) of hsp65 gene.20 These strains were randomly selected from a collection of previously described biofilm-forming strains.16M. fortuitum ATCC 6841T, M. peregrinum ATCC 14467T, M. abscessus DSM 44196T, M. chelonae ATCC 35752T, M. mucogenicum DSM 44124, and M. mageritense ATCC 700351T were included in the studies as controls.
Antimicrobial susceptibilityThe strains were tested for amikacin (SIGMA, St. Louis, Missouri, USA), clarithromycin (Abbott Park, Illinois, USA), and ciprofloxacin (SIGMA, St. Louis, Missouri, USA) susceptibility using the microdilution reference technique as previously described.21 These antibiotics were selected because they are the ones most commonly used to treat NPRGM infections.
Antimicrobial susceptibility of biofilmsWe obtain mature biofilms on sterile polycarbonate disks (ALBET-Hahnemuehle, Dassel, Germany) as described previously,22 except that we used Tryptic soy-5% sheep blood agar plates instead of Müller-Hinton agar plates. After 5 days of incubation at 37°C, disks were removed using sterile forceps and placed onto Müller-Hinton agar plates containing 50μg/ml of amikacin, clarithromycin, and ciprofloxacin, respectively. After 24hours at room temperature, disks were removed and introduced into 5ml of sterile distilled water. We then dislodged the biofilm by vortexing for 1minute and sonication (Ultrasons-H 3000840; J.P Selecta, Abrera, Spain) for 5minutes. The number of colony forming units (CFU) of viable bacteria was quantified by streaking serial dilutions from the sonicate on Tryptic soy-5% sheep blood agar plates and incubating them for 5 days at 37°C. All the strains were tested in triplicate.
Antibiotic permeability assayWe used the method described by Anderl et al.22 with modifications. A 0.5 McFarland suspension of each strain was prepared in sterile PBS using glass beads and vortexing. Ten μl of each suspension were inoculated onto a sterile polycarbonate disk (ALBET-Hahnemuehle, Dassel, Germany), previously placed on Muller-Hinton agar plates (bioMérieux, Marcy-l’Etoile, France) and were incubated at 37°C for 7 days. After this period, another polycarbonate disk was placed onto the developed biofilm and a 6-mm diameter sterile paper disk with a previously known amount of antibiotic (15μg for clarithromycin and ciprofloxacin, and 30μg for amikacin) was moistened with 15μl of Muller-Hinton broth (bioMérieux, Marcy-l’Etoile, France) and placed on top of the membrane-biofilm-membrane “sandwich”. The upper disks were removed after 3, 6, 24 and 48hours at room temperature and the remaining antibiotic concentration was quantified by a disk-plate assay on Müller-Hinton agar plates (bioMérieux, Marcy l’Etoile, France) using Kocuria rizophyla ATCC 9341. Previously, we established a relationship between the inhibition diameter of K. rhizophila ATCC 9341 and different concentrations of antibiotics. The measurements of the inhibition diameter were taken with a digital calliper after 24 hour incubation at 37°C in normal atmosphere.
We used identical “sandwich” structures of polycarbonate disks without any biofilm as positive controls for antimicrobial permeability, and a Parafilm© membrane instead the biofilm as a control for non-permeability (negative control). The negative control measurements were used to calculate the percentage of antibiotic concentration in the disk in order to avoid false results due to antibiotic degradation.
All experiments (including standard bioassays) were performed in triplicate.
Statistical analysisTo analyze the antimicrobial resistance of biofilms, we calculated the percentage of viable bacteria recovered after exposure, using the mean of controls without antibiotics as the 100% value.
We checked the normality of the series using the Bartlett test. For comparison of the mean values, ANOVA and Mann-Whitney/Wilcoxon tests were performed when indicated. All calculations were performed with the EPI-INFO 3.4.1 software (Center for Disease Control and Prevention, Atlanta, USA).
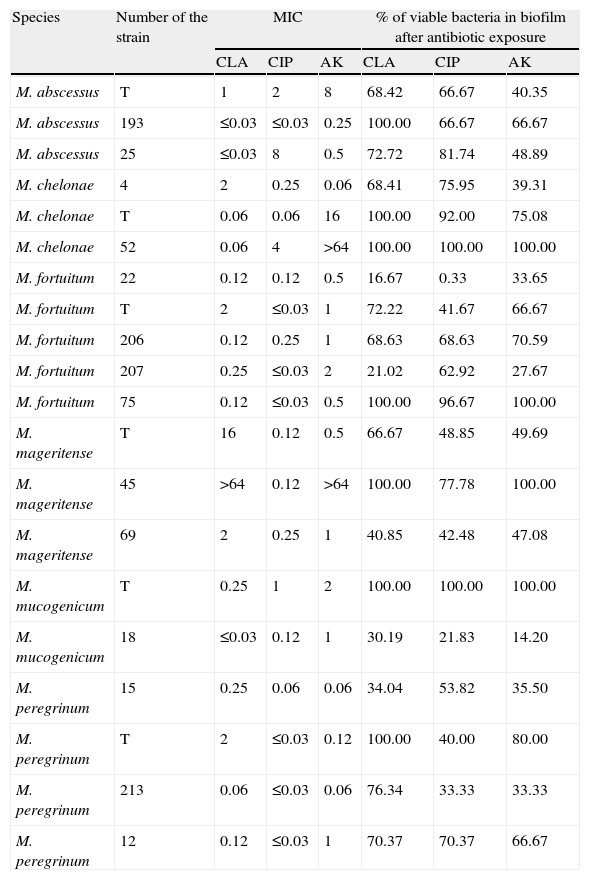
ResultsThe number of viable bacteria recovered from the biofilms after antimicrobial exposure ranged between 3.2 × 106 CFU/ml and 4.0 × 1014 CFU/ml (Table 1).
Results obtained with the different strains tested.
| Species | Number of the strain | MIC | % of viable bacteria in biofilm after antibiotic exposure | ||||
| CLA | CIP | AK | CLA | CIP | AK | ||
| M. abscessus | T | 1 | 2 | 8 | 68.42 | 66.67 | 40.35 |
| M. abscessus | 193 | ≤0.03 | ≤0.03 | 0.25 | 100.00 | 66.67 | 66.67 |
| M. abscessus | 25 | ≤0.03 | 8 | 0.5 | 72.72 | 81.74 | 48.89 |
| M. chelonae | 4 | 2 | 0.25 | 0.06 | 68.41 | 75.95 | 39.31 |
| M. chelonae | T | 0.06 | 0.06 | 16 | 100.00 | 92.00 | 75.08 |
| M. chelonae | 52 | 0.06 | 4 | >64 | 100.00 | 100.00 | 100.00 |
| M. fortuitum | 22 | 0.12 | 0.12 | 0.5 | 16.67 | 0.33 | 33.65 |
| M. fortuitum | T | 2 | ≤0.03 | 1 | 72.22 | 41.67 | 66.67 |
| M. fortuitum | 206 | 0.12 | 0.25 | 1 | 68.63 | 68.63 | 70.59 |
| M. fortuitum | 207 | 0.25 | ≤0.03 | 2 | 21.02 | 62.92 | 27.67 |
| M. fortuitum | 75 | 0.12 | ≤0.03 | 0.5 | 100.00 | 96.67 | 100.00 |
| M. mageritense | T | 16 | 0.12 | 0.5 | 66.67 | 48.85 | 49.69 |
| M. mageritense | 45 | >64 | 0.12 | >64 | 100.00 | 77.78 | 100.00 |
| M. mageritense | 69 | 2 | 0.25 | 1 | 40.85 | 42.48 | 47.08 |
| M. mucogenicum | T | 0.25 | 1 | 2 | 100.00 | 100.00 | 100.00 |
| M. mucogenicum | 18 | ≤0.03 | 0.12 | 1 | 30.19 | 21.83 | 14.20 |
| M. peregrinum | 15 | 0.25 | 0.06 | 0.06 | 34.04 | 53.82 | 35.50 |
| M. peregrinum | T | 2 | ≤0.03 | 0.12 | 100.00 | 40.00 | 80.00 |
| M. peregrinum | 213 | 0.06 | ≤0.03 | 0.06 | 76.34 | 33.33 | 33.33 |
| M. peregrinum | 12 | 0.12 | ≤0.03 | 1 | 70.37 | 70.37 | 66.67 |
CLA: Clarithromycin; CIP: Ciprofloxacin, AK: Amikacin; T: Type strain.
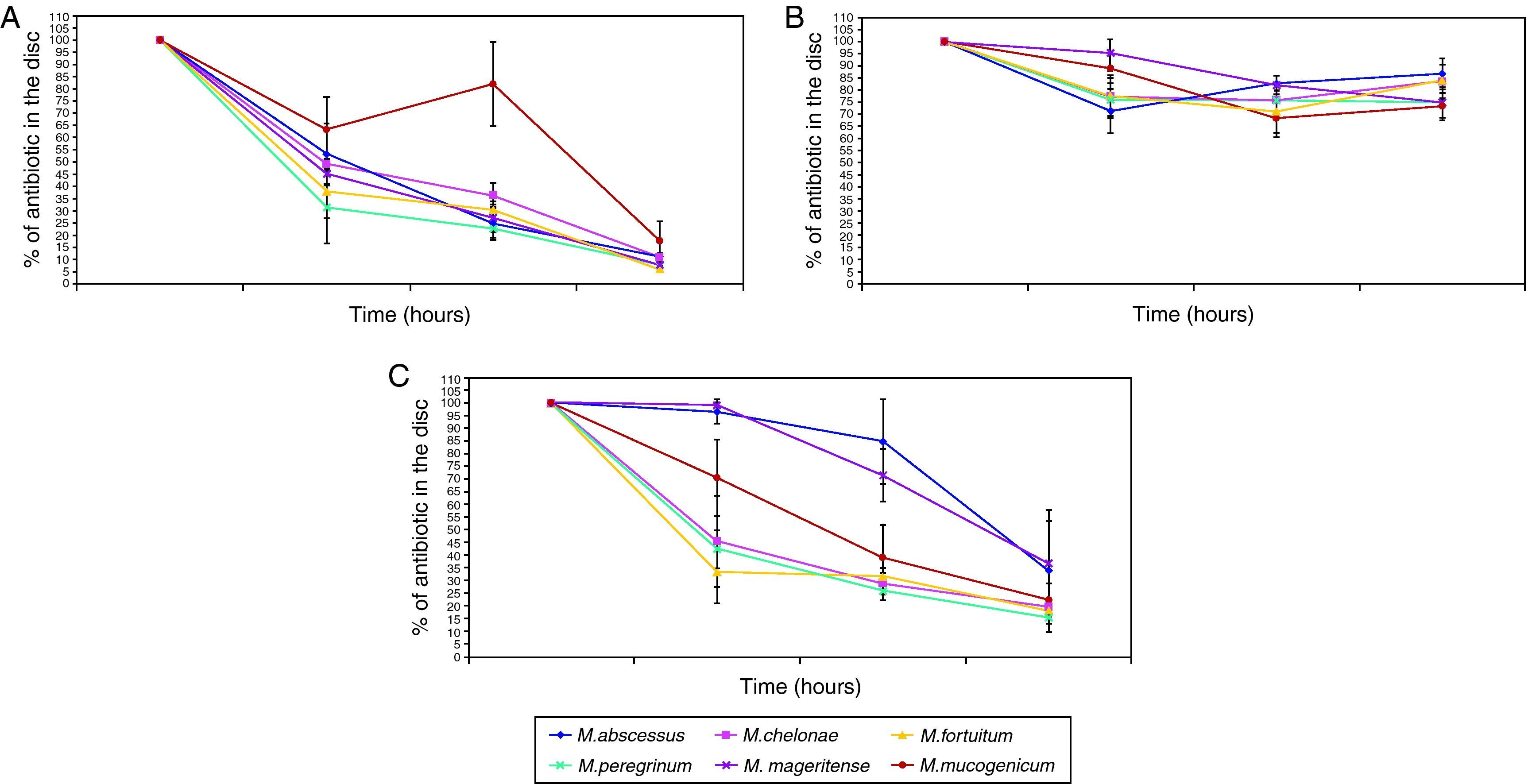
All antibiotic-treated biofilms showed a decrease in the percentage of viable bacteria, and no differences could be found between the different antibiotics tested. If we use the criterion for bactericidal concentration of biofilm23 previously described, all biofilms produced would be considered resistant against all tested antibiotics. However, despite there being no overall differences observed between mycobacteria species and different antibiotics, a high intraspecies variation was observed (Table 1). No studies regarding antimicrobial susceptibility of released viable bacteria or development of antimicrobial resistance among them were performed.
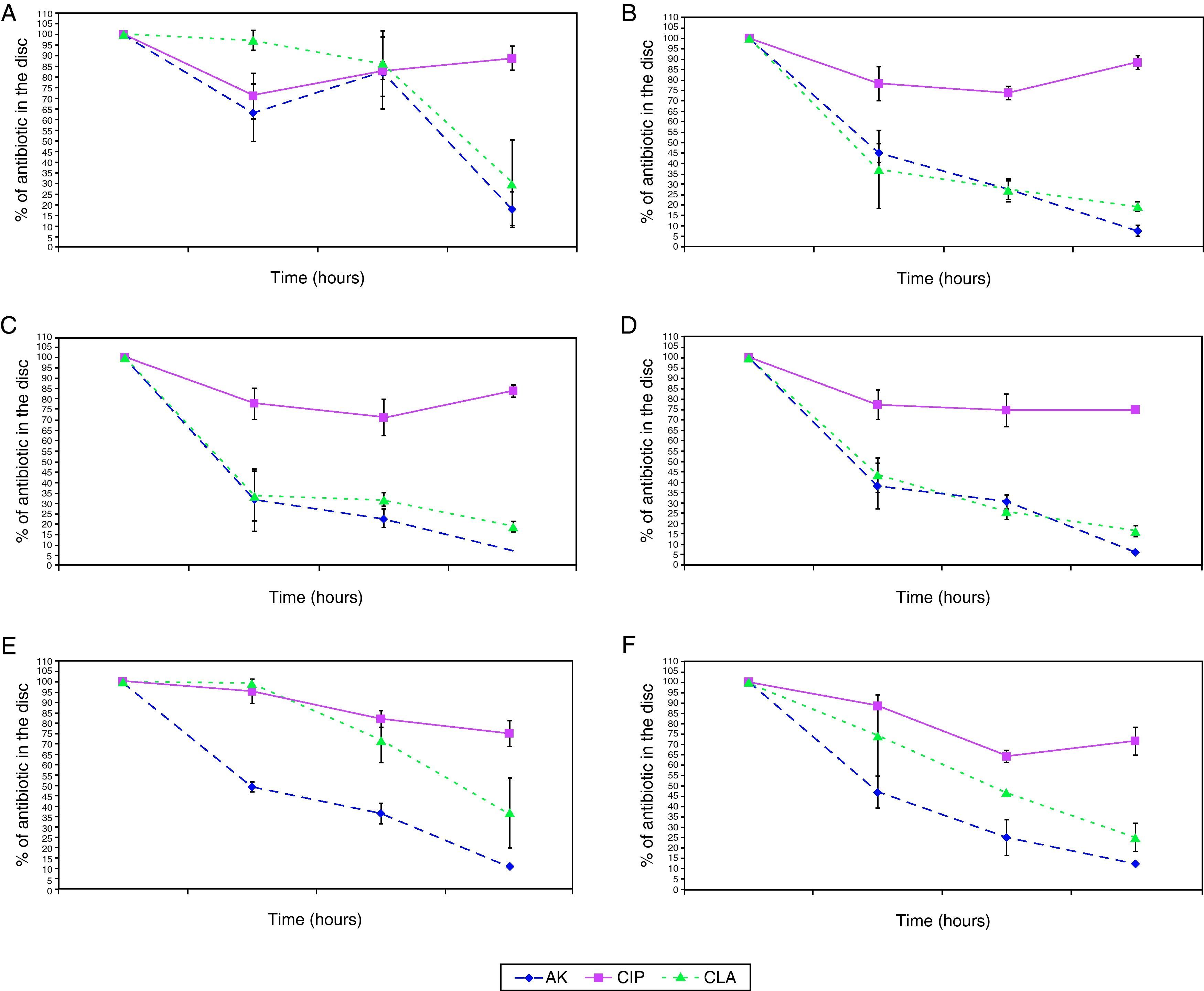
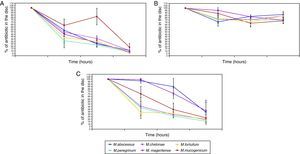
The penetration curves in the biofilm for the three tested antibiotics were different (Figures 1 and 2). Amikacin was the antibiotic with the highest penetrability into biofilm, showing the smallest percentage of antibiotic in the disk at 24hours (10.56%). Clarithromycin showed an intermediate penetration (23.96%) and Ciprofloxacin had a poor penetration in the biofilm, with a mean amount of 80.39% at 24hours in the disk. This pattern was shared for all strains, in spite of the in vitro susceptibility differences of biofilms.
Penetration of antibiotics in the different species. Percentage of antibiotic that remains in the disk after 0, 3, 6, and 24hours. 1A: M. abscessus, 1B: M. chelonae, 1C: M. fortuitum, 1D: M. peregrinum, 1E: M. mageritense, 1F: M. mucogenicum.
Footnote: AK: Amikacin, CIP: Ciprofloxacin, CLA: Clarithromycin.
When we analysed the antimicrobial penetration with time of exposure, we observed a rapid penetration of amikacin and clarithromycin for all species, except for M. abscessus and M. mageritense. For clarithromycin, while the majority of species showed percentages of antibiotic remaining after 6hours of exposure between 26.2-31.8%, M. mucogenicum showed 39.3%, M. mageritense 71.6% and M. abscessus 84.9%. For amikacin, M. abscessus showed a percentage of 67.2% of antibiotic after 6hours of exposure, in comparison with 22.8-36.4% for the rest of the remaining species. These data suggest a more rapid penetration of antibiotic in some species, and a slower one in others. Such differences did not appear for ciprofloxacin.
DiscussionOne of the reports that have evaluated the antimicrobial effect against biofilms formed by different species of mycobacteria showed an antimicrobial resistance increase of Mycobacterium avium biofilms against clarithromycin and rifampin, although their authors stated that these results were probably not due to permeability problems as the biofilm was extremely immature.24 A more recent study reported the resistance of Mycobacterium tuberculosis biofilms against isoniazid and rifampin, suggesting that metabolic changes were probably responsible.25 Among rapidly growing mycobacteria, some studies have shown the resistance to a broad spectrum of biocides.17,26 A study of Mycobacterium smegmatis biofilm reported its isoniazid resistance and ruled out the slower biofilm growth as cause, with the authors suggesting that permeability features and/or other unknown mechanisms were responsible.18 Another report showed that M. abscessus biofilms were resistant to cefoxitin, amikacin and clarithromycin, the nutrient limitations and a latent metabolic state being proposed as responsible.19 Recent studies have shown that extracellular matrix in mycobacterial biofilm is formed mainly by lipids, including mycolic acids, but the actual role of these components in antimicrobial penetration is unknown.27,28
Our study is the first to evaluate biofilm resistance for different clinical species of mycobacteria. Of interest, despite the lack of differences observed in the percentages of viable bacteria obtained after exposure to antibiotics, important variations in the individual susceptibility of different strains biofilms were reported, as well as the individual variations previously observed for adherence,29 the first crucial step for biofilm development. Due to these observation, we studied one of the mechanisms claimed as important in biofilm resistance, the antibiotic penetration through biofilm.
Until now, no studies have analyzed the antimicrobial permeability for biofilms of different species of mycobacteria. In our analysis, amikacin showed better penetration than clarithromycin, but no statistically significant differences could be found between the activities of both antibiotics. However, a different pattern of penetration could be observed for ciprofloxacin, showing the minimal penetration through biofilm for all strains. These results could indicate, that despite of differences in antibiotic penetration, this phenomenon could have a minimal role (if any) in antibiotic resistance of NPRGM biofilms, because mature biofilms of all strains analyzed are resistant to tested antibiotics independently of the antibiotic penetration pattern.
These results are different to those obtained in other studies, where ciprofloxacin seemed to penetrate the biofilm formed by Pseudomonas aeruginosa,30Klebsiella pneumoniae22 or Staphylococcus epidermidis31 better than other antibiotics. In the study performed by Walters et al.30 the penetration of ciprofloxacin through a P. aeruginosa biofilm was very much higher than tobramycin. In our study, the comparison of ciprofloxacin and amikacin (the aminoglycoside used in the treatment of NPRGM infections) showed that the penetration of ciprofloxacin is very much lower than amikacin. Differences in the composition of the extracellular matrix could be the reason for these differences, as the matrix mainly consists of different polysaccharides in P. aeruginosa32, while recent studies have showed that mycobacteria produce a lipid-rich matrix with a high content of mycolic acids25 and glycopeptidolipids.33
In contrast to others studies, we have analyzed not only collection strains, but also clinical isolates. Genetic differences between wild strains and laboratory ones have been reported,34 and these differences should be considered in the evaluation of antibiotic resistance of biofilms. In our study, no differences were detected in antimicrobial permeability in biofilms of different strains, although they do have different antimicrobial susceptibility (Table 1). These observations could indicate that antimicrobial permeability features are not species-dependent nor related to drug resistance.
It would be of interest to explore the effect of a combination of antibiotics on mycobacterial biofilms, especially because therapy of the infections caused by these organisms in usually combined. Moreover, recent data suggest that macrolides have a quorum sensing inhibition effect in biofilms produced by Pseudomonas aeruginosa.28 Further studies are necessary to evaluate this effect of macrolides in NPRGM biofilms, both alone or combined with other antimicrobials. More studies are also needed to confirm whether NPRGM biofilms are resistant to other antimicrobials.
In conclusion, NPRGM biofilms showed drug resistance against antibiotics that are commonly used for the treatment of infections caused by these organisms. We have detected differences in antibiotic penetration through biofilms with an important permeability barrier for ciprofloxacin. However, as there were no differences observed in the resistance for the tested antibiotics, it is likely that this mechanism could have a low importance for antimicrobial resistance of biofilms. Other mechanisms, such as metabolic latency, are probably more important in explaining the antimicrobial resistance of NPRGM biofilm.
Conflict of interestThe authors have no conflict of interest to declare.
This work was funded by grants from Capio Research Foundation (2006-1159 and 2007-1457), Comunidad de Madrid (S-0505/MAT/000324) and CONSOLIDER 2010 program (FUNCOAT-CSD2008-00023). Nieves Z. Martín-de-Hijas and Diana Molina-Manso were funded by the Fundación Conchita Rábago de Jiménez Díaz. Noelia Alonso-Rodriguez was funded by the Comunidad de Madrid.
No conflict of interest for all the authors.