Colistin resistance can occur by chromosomal mutations and by acquisition of plasmid-carrying determinants, mainly mcr-1. In the recent years, we have observed the outburst of this resistance gene in our region. Due to the risk of the rapid dissemination of mcr-1, this finding has worried and alerted different actors from the health field and has become one of the most prolific topics. Our review compiles available reports of well-documented mcr-1-positive strains of Enterobacteriaceae, obtained from different samples in Argentina and other countries of Latin America. Furthermore, it addresses the association of mcr-1 with ESBL resistance markers and outlines the platforms involved in their dissemination.
La resistencia a la colistina puede ocurrir por mutaciones cromosómicas o por la adquisición de determinantes localizados en plásmidos, el principal es mcr-1. En los últimos años hemos observado la explosiva aparición de este gen de resistencia en nuestra región. Debido al riesgo que implica la rápida diseminación de mcr-1, este hallazgo ha preocupado y alertado a los diferentes actores del área de la salud, y se ha convertido en uno de los temas de investigación más importantes. La presente revisión compila los informes de aislamientos portadores de mcr-1 debidamente documentados en Enterobacteriaceae, obtenidos de diferentes muestras en Argentina y otros países de América Latina. Además, aborda la asociación de mcr-1 con otros marcadores de resistencia, como las BLEE, y describe las plataformas involucradas en su diseminación.
Polymyxins (colistin and polymyxin B) are polycationic peptides, which act by targeting the negative charges in bacterial lipopolysaccharide (LPS), the complex lipoglycan that makes up most of the gram-negative outer membrane. Colistin (Polymyxin E) is one of the few cationic antimicrobial peptides commercialized in both human and veterinary medicine. Since its introduction in the 1950s, colistin (COL) has been used mainly as a topical treatment in human medicine owing to its nephrotoxicity and neurotoxicity when given systemically. There are two commercially available forms: colistin sulphate (CS) and colistin methanesulphonate sodium (CMS), the latter being less toxic than colistin when administered parenterally and consequently being the most used both in parenteral and inhalational formulations. In aqueous solutions, CMS undergoes in vivo conversion9. On the other hand, CS is the only form of COL approved in pig production in some countries for the control of Enterobacteriaceae infections, particularly for those caused by Escherichia coli. However, COL has been massively used at sub-inhibitory concentrations as growth promoters for decades to improve weight gain and therefore to maximize feed conversion efficiency in animal production22. Noteworthy, the last survey conducted by the World Organization for Animal Health (OIE) in the year 2012 revealed that only 10 countries reported the use of COL as growth promoter42. Interestingly, in the late 2000s and after decades of COL use in swine, several studies began to report a significant COL resistance rate in Enterobacteriaceae1,4,23,33,49,55. The increasing incidence of infections caused by multidrug resistant gram-negative bacteria, especially the ones caused by carbapenemase-producing bacilli in the last years, has led to the use of polymyxins as one of the last resort treatments49.
Until very recently, COL resistance occurred by chromosomal mutations only, and therefore, this resistance phenotype was spread exclusively via clonal transmission of resistant isolates. However, two years ago Liu et al. reported the emergence of a plasmid-mediated colistin resistance determinant, mcr-139. Due to the risk of its rapid dissemination, this finding has worried and alerted different actors from the health field, becoming one of the most prolific topics.
Acquired resistance to colistinAcquired resistance to colistin in Enterobacteriaceae was described many years ago45,64, but in the last years colistin-resistant K. pneumoniae isolates were more frequently reported worldwide10,11,32,52, among others. In Latin America there are reports from Brazil, Ecuador, Venezuela and Colombia18,27,50,56,58.
Acquired resistance to COL in Enterobacteriaceae was first documented in Argentina in the last years together with the emergence of KPC-producing isolates, probably derived from the use of this antimicrobial agent in the therapy of infections by these multidrug-resistant K. pneumoniae isolates. This resistance is believed to have occurred by selection of resistant subpopulations in the epidemic clone belonging to ST258. However, it has also been observed in non KPC-producing K. pneumoniae isolates probably as a result of clonal spread43. The last report of the PAHO (Panamerican Health Organization) from the year 2015 informed a COL resistance rate of 10.3% in K. pneumoniae isolates from hospitalized patients in Argentina, whereas in Escherichia coli and Enterobacter cloacae the percentage only reached 2.1%51. Similar results were obtained at the University Hospital José de San Martín, where COL resistance in K. pneumoniae in the year 2010 was 2% and 3%, 3.6%, 3.8%, 7%, 12% and 8%, respectively, in the subsequent years (unpublished results). With regard to E. coli isolates, COL resistance emerged in the last two years, where an important increase from 0.2% to 1.3% was observed, mostly as a result of plasmid-mediated resistance44. On the other hand, acquired resistance to COL remains very low in Acinetobacter baumannii (0.2%) and Pseudomonas aeruginosa (2.5%) isolates51.
Chromosomal resistance: different pathwaysBacteria often use different strategies to protect themselves from exposure to cationic antimicrobial peptides such as colistin. Such strategies include modifications of their lipopolysaccharides (LPSs), which are involved in the initial step of interaction between polymyxins and bacteria8,47,48. These alterations can be achieved by several covalent changes in lipid A (also called endotoxin molecule) of LPS. Among them, the cationic substitution of the phosphate group by the addition of 4-amino-4-deoxy-l-arabinose (L-Ara4N) and phosphoethanolamine (PEtN) are the most common LPS modifications. L-Ara4N modification is more effective than PEtN modification because the decrease in the net charge leading to colistin resistance is more effective. The synthesis and transfer of L-Ara4N and PEtN is mediated by an operon regulated by the activation of two-component systems (TCSs). PmrA/PmrB is the main system contributing to polymyxin resistance (by environmental stimuli, and specific mutations within the TCS), although in some gram-negative species is known to occur via PhoP/PhoQ8,48.
Other modifications associated to chromosomal polymyxin resistance have been described, such as the reduction of acyl groups mediated by an lpxR-like deacylation and hydroxylation of lipid A48. Moreover, it was reported that K. pneumoniae uses alternative strategies such as participation of an efflux pump or capsule formation48.
The plasmid-mediated resistance gene mcr-1In 2015, the emergence of a plasmid-mediated colistin resistance marker, named mcr-1, was reported in China39. Since then, numerous reports of the mcr-1 gene have been documented in Enterobacteriaceae cultured from food, animals, environment and humans worldwide, most of them detected in over 20 countries within 3 months of its first report8.
The mcr-1 gene encodes a PEtN transferase leading to the addition of PEtN to the lipid A. An in silico analysis of the amino acid sequence of this protein revealed that it is strongly related to enzymes present in gram-negative bacteria, some of which are intrinsically resistant to polymyxin8,19,48,52. This gene was detected mainly in E. coli and to a lesser extent, in other bacteria such as K. pneumoniae. To date, several variants of mcr-1 have been reported in the literature (mcr-1.2 to mcr-1.8, and mcr-1.10)2,20,40,66,69 or are deposited in Genbank (variants mcr-1.9, mcr-1.11, mcr-1.12, and mcr-1.13). These variants have one amino acid change each and exhibit a similar effect on colistin resistance compared with mcr-1.
Very recently, four additional mcr determinants have been reported, mcr-2, mcr-3, mcr-4 and mcr-5. The mcr-2 gene was identified in an E. coli isolated from pigs and cattle in Belgium68. This marker was also recovered from human samples in Switzerland37. Phylogenetic analysis of mcr-2, which was also described as a PEtN transferase, showed 80.6% amino acid identity with mcr-1 and provided strong evidence that this protein may have originated from Moraxella catarrhalis68. The mcr-3 gene was found in E. coli isolated from pigs in Malaysia, in a clinical sample of K. pneumoniae isolated from Thailand, and a clinical sample of Salmonella Typhimurium from the United States70. Sequence analysis showed that mcr-3 is closely related to PEtN from Aeromonas spp. The mcr-4 gene was found in Salmonella Typhimurium and E. coli from human and food-producing animals in Europe13. MCR-4 has 34.0%, 35.0% and 49.0% amino acid sequence identity to MCR-1, MCR-2 and MCR-3, respectively. It has been proposed that mcr-4 may have emerged from a Shewanella species, a bacterium commonly present in aquatic niches. Lastly, mcr-5 was initially found in 14 Salmonella Paratyphi B dTa+ isolates12. MCR-5 shares 36.11% protein identity with MCR-1, 35.29% with MCR-2, 34.72% with MCR-3 and 33.71% with MCR-4. Sequence analysis revealed that mcr-5 might have been transferred from the environmental rod Cupriavidus gilardii.
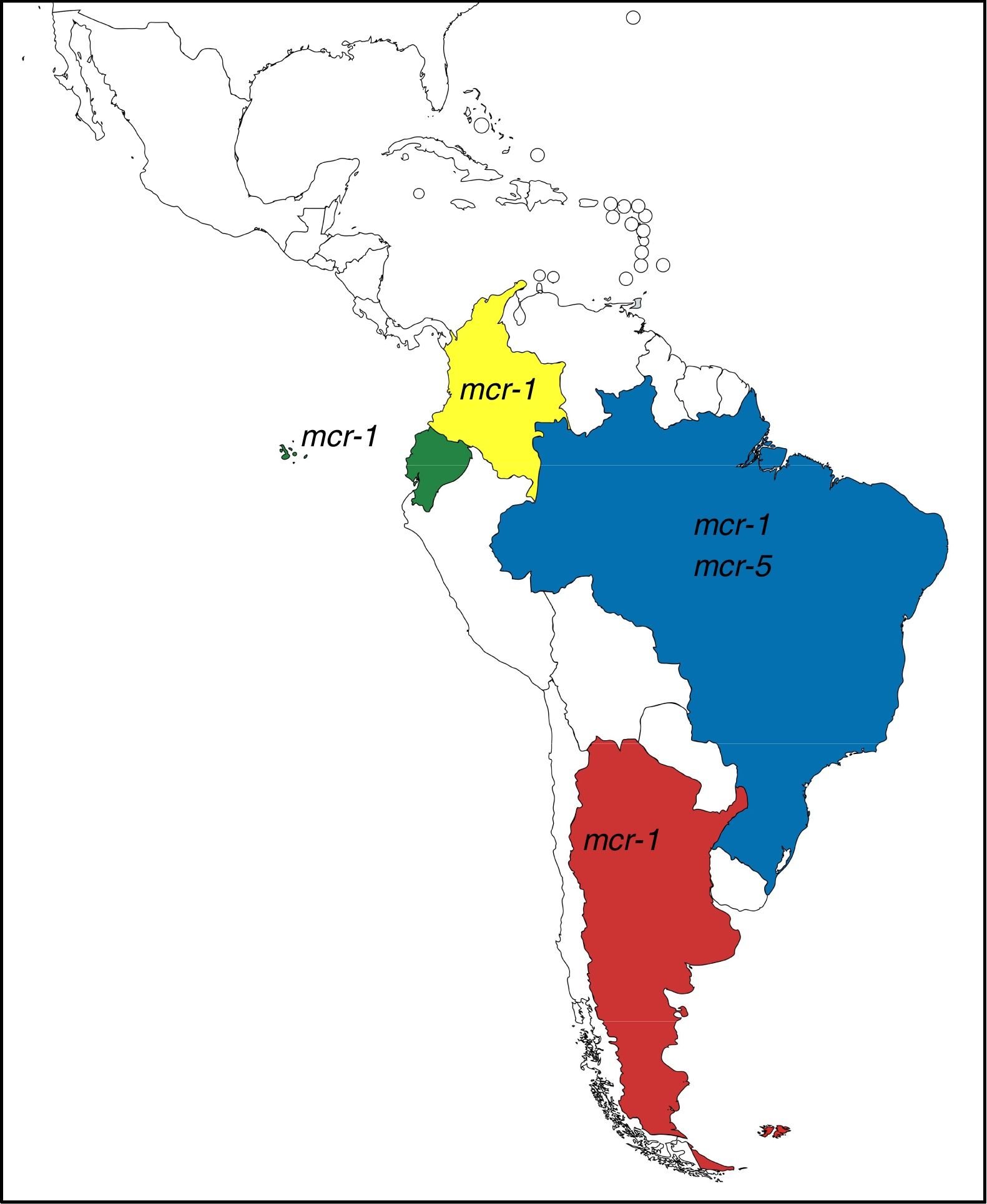
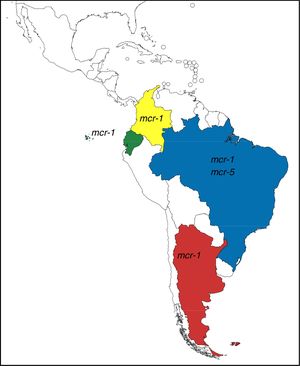
These genes have completely changed the scenario of colistin resistance since they have become a potential threat to public health. In Argentina and other countries of Latin America, mcr-1 occurrence is currently low, however, it is possible to observe its rapid increase (Table 1). Moreover, mcr-5 has been recently found in a colistin-resistant E. coli isolate from Brazil (NZ_PDMU00000000) (Fig. 1).
mcr-1-positive isolates reported in Latin America
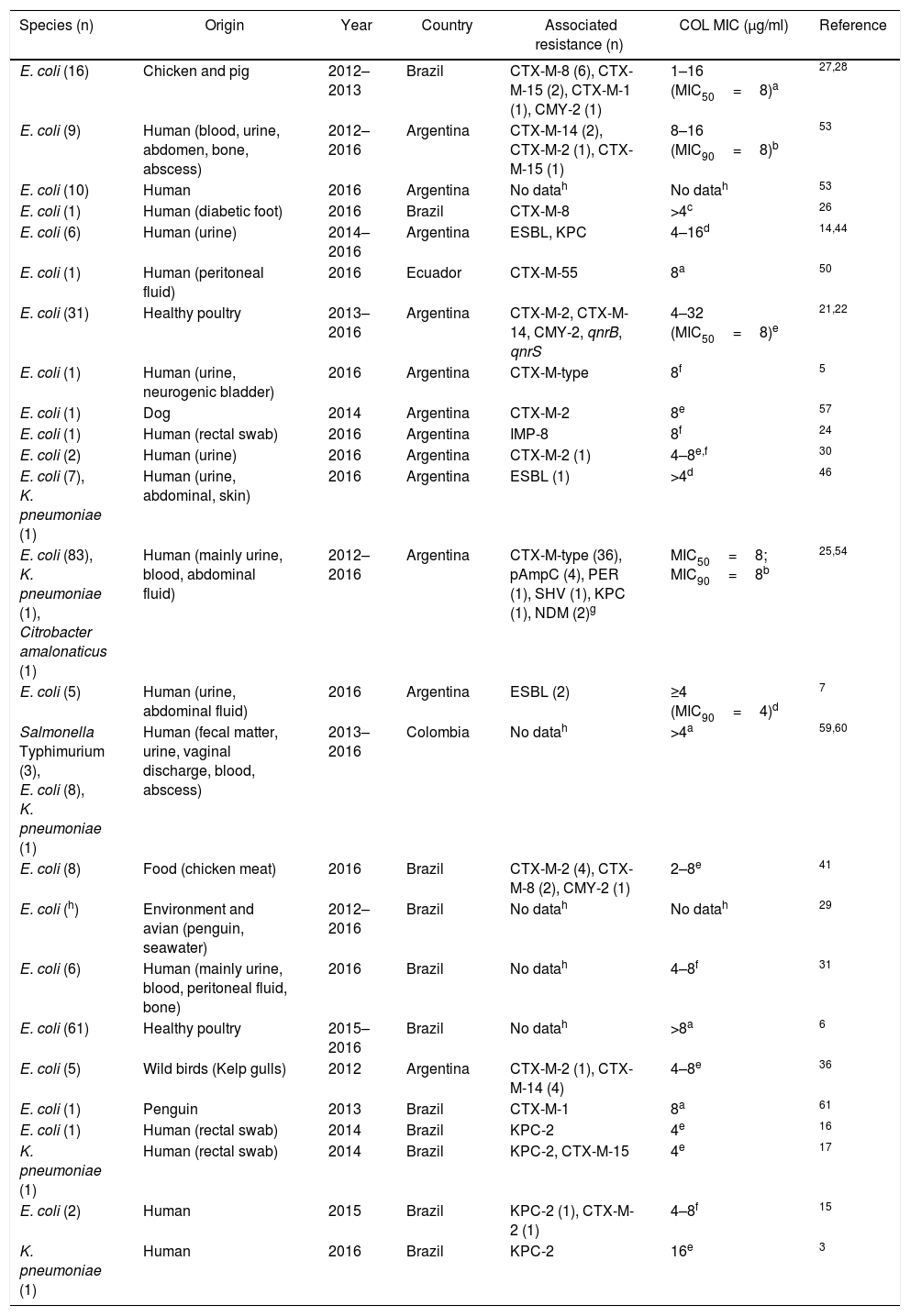
| Species (n) | Origin | Year | Country | Associated resistance (n) | COL MIC (μg/ml) | Reference |
|---|---|---|---|---|---|---|
| E. coli (16) | Chicken and pig | 2012–2013 | Brazil | CTX-M-8 (6), CTX-M-15 (2), CTX-M-1 (1), CMY-2 (1) | 1–16 (MIC50=8)a | 27,28 |
| E. coli (9) | Human (blood, urine, abdomen, bone, abscess) | 2012–2016 | Argentina | CTX-M-14 (2), CTX-M-2 (1), CTX-M-15 (1) | 8–16 (MIC90=8)b | 53 |
| E. coli (10) | Human | 2016 | Argentina | No datah | No datah | 53 |
| E. coli (1) | Human (diabetic foot) | 2016 | Brazil | CTX-M-8 | >4c | 26 |
| E. coli (6) | Human (urine) | 2014–2016 | Argentina | ESBL, KPC | 4–16d | 14,44 |
| E. coli (1) | Human (peritoneal fluid) | 2016 | Ecuador | CTX-M-55 | 8a | 50 |
| E. coli (31) | Healthy poultry | 2013–2016 | Argentina | CTX-M-2, CTX-M-14, CMY-2, qnrB, qnrS | 4–32 (MIC50=8)e | 21,22 |
| E. coli (1) | Human (urine, neurogenic bladder) | 2016 | Argentina | CTX-M-type | 8f | 5 |
| E. coli (1) | Dog | 2014 | Argentina | CTX-M-2 | 8e | 57 |
| E. coli (1) | Human (rectal swab) | 2016 | Argentina | IMP-8 | 8f | 24 |
| E. coli (2) | Human (urine) | 2016 | Argentina | CTX-M-2 (1) | 4–8e,f | 30 |
| E. coli (7), K. pneumoniae (1) | Human (urine, abdominal, skin) | 2016 | Argentina | ESBL (1) | >4d | 46 |
| E. coli (83), K. pneumoniae (1), Citrobacter amalonaticus (1) | Human (mainly urine, blood, abdominal fluid) | 2012–2016 | Argentina | CTX-M-type (36), pAmpC (4), PER (1), SHV (1), KPC (1), NDM (2)g | MIC50=8; MIC90=8b | 25,54 |
| E. coli (5) | Human (urine, abdominal fluid) | 2016 | Argentina | ESBL (2) | ≥4 (MIC90=4)d | 7 |
| Salmonella Typhimurium (3), E. coli (8), K. pneumoniae (1) | Human (fecal matter, urine, vaginal discharge, blood, abscess) | 2013–2016 | Colombia | No datah | >4a | 59,60 |
| E. coli (8) | Food (chicken meat) | 2016 | Brazil | CTX-M-2 (4), CTX-M-8 (2), CMY-2 (1) | 2–8e | 41 |
| E. coli (h) | Environment and avian (penguin, seawater) | 2012–2016 | Brazil | No datah | No datah | 29 |
| E. coli (6) | Human (mainly urine, blood, peritoneal fluid, bone) | 2016 | Brazil | No datah | 4–8f | 31 |
| E. coli (61) | Healthy poultry | 2015–2016 | Brazil | No datah | >8a | 6 |
| E. coli (5) | Wild birds (Kelp gulls) | 2012 | Argentina | CTX-M-2 (1), CTX-M-14 (4) | 4–8e | 36 |
| E. coli (1) | Penguin | 2013 | Brazil | CTX-M-1 | 8a | 61 |
| E. coli (1) | Human (rectal swab) | 2014 | Brazil | KPC-2 | 4e | 16 |
| K. pneumoniae (1) | Human (rectal swab) | 2014 | Brazil | KPC-2, CTX-M-15 | 4e | 17 |
| E. coli (2) | Human | 2015 | Brazil | KPC-2 (1), CTX-M-2 (1) | 4–8f | 15 |
| K. pneumoniae (1) | Human | 2016 | Brazil | KPC-2 | 16e | 3 |
Susceptibility to colistin was evaluated by: (a) unspecified method, (b) agar dilution, (c) MicroScan system (Beckman Coulter), (d) Phoenix (Becton, Dickinson), (e) microdilution method, (f) Vitek-2, (g) NDM was detected in C. amalonaticus, (h) no data described in the Abstract.
Furthermore, there is a clear association between this determinant and the different genes encoding β-lactamases, particularly those from the blaCTX-M family. It is worth mentioning that mcr-1 can be found in isolates collected not only from human and animal clinical samples but also from different sources such as pigs, wild birds, penguins, poultry production and environmental niches (Table 1). Among these isolates, E. coli was the most common species reported in Brazil and Argentina.
The coexistence of mcr-1 and carbapenem resistance determinants (such as KPC-2, NDM and IMP-8 carbapenemases) in human samples is of great concern (Table 1). These findings highlight that Enterobacteriaceae isolates carrying both mcr-1 and carbapenemase-encoding genes may emerge as a serious threat to antimicrobial therapy.
Genetic environment and dissemination of the mcr genePrevious studies have shown that mcr genes are commonly located in self-transferable plasmids, which are responsible for inter-species dissemination worldwide. Plasmid incompatibility groups IncI2, HI1, HI2, P, X1/X2, X3/X4, X4, FI, FIP and FII have been reported as carriers of the different mcr variants12,14,18,23,26,34,39–41,52,58,68,70. Furthermore, a hybrid plasmid, identified as IncI2/IncFIB also harbored this resistance gene67. In South America only a few reports identified the plasmids responsible for mcr spread. To date, only plasmids from incompatibility groups X4 and I2 carry the mcr gene in our region3,21,41,66. An initial analysis of the plasmid backbone suggests that these elements are highly similar to those reported in China, Europe or North America.
A common feature of mcr-1 variants is their genetic environment. This gene is commonly adjacent to the pap2 gene. Both genes can be found either surrounded by two copies of insertion sequence ISApl1, yielding a composite transposon, downstream to a single copy of ISApl1 or in the absence of this element62. It has been proposed that the presence or absence of this IS correlates with the adaptation of mcr-1 to a new host. A composite transposon indicates a recent acquisition of this marker whereas the absence of ISApl1 or the presence of a single copy suggests that it has been already adapted. Since all three genetic environments have been found in different plasmids, it has been hypothesized that independent recombination events have led to the adaptation of the mcr-pap2 segment to specific incompatibility groups, which are in turn responsible for dissemination of polymyxin resistance. However, a recent report demonstrated that the composite transposon Tn6330, which has two ISApl1 surrounding mcr-1, can translocate to other plasmids35. Therefore, the possibility of a wider dissemination to plasmids from other incompatibility groups should be considered. It is worth noting that a study on ISApl1 function showed that when this sequence is active it is detrimental to the host, which leads to its loss63. Indirectly, this phenomenon might control the dissemination of mcr-1.
In 2016 Xavier et al. identified the first mcr-2 gene in an IncX4 plasmid from bovine and porcine E. coli68. Further analysis showed that mcr-2 is located at the same site than mcr-1 when found in the IncX4 plasmid. Despite this fact, mcr-2 was found surrounded by two copies of the insertion sequence IS1595 allowing this DNA to generate circular intermediates65. This finding reveals the potentiality of mcr-2 to disseminate to other plasmids and therefore, to other organisms. More recently, novel mcr determinants have been described. The mcr-3 gene has been found in plasmids from the IncHI2 group harbored in Enterobactericeae as well as in the chromosome of Aeromonas veronii38,70. When located in plasmids, mcr-3 is adjacent to the truncated transposon element TnAs270, a mobile element probably responsible for its spread. On the other hand, mcr-4 and mcr-5 genes were found in small non-self-transferable plasmids from the ColE family12,13. Furthermore, the mcr-5 gene is located in a Tn3-family transposon.
Concluding remarksThe data summarized herein show the rapid spread of mcr-1 in Latin America. To the best of our knowledge, mcr-2, mcr-3 and mcr-4 determinants have not been found in our region yet.
The mcr-1 marker has been found in a wide variety of ecological niches, which represents a troublesome scenario for public health. In the absence of new antimicrobial agents against multidrug-resistant gram-negative pathogens, the effect of plasmid-mediated colistin resistance on human health should not be underestimated.
There is an urgent need for surveillance and molecular epidemiological studies in both human and veterinary medicine addressing the dissemination of mcr-1 among bacterial isolates. The fact that most mcr genes were found in other regions raises the question of whether a unique variant is locally disseminated or if the other variants were accurately sought.
Furthermore, we suggest a re-evaluation of the antimicrobial agents used in animal feeding in order to reduce the potential risk to pandrug resistance.
FundingThis work was supported by ANPCyT PICT 2013-1978, UBACyT 20020130100167BA and ANPCyT PICT 2015-1925.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors would like to thank Famiglietti A, Rodriguez CH and Gutkind G for their invaluable cooperation.